Figure 7.
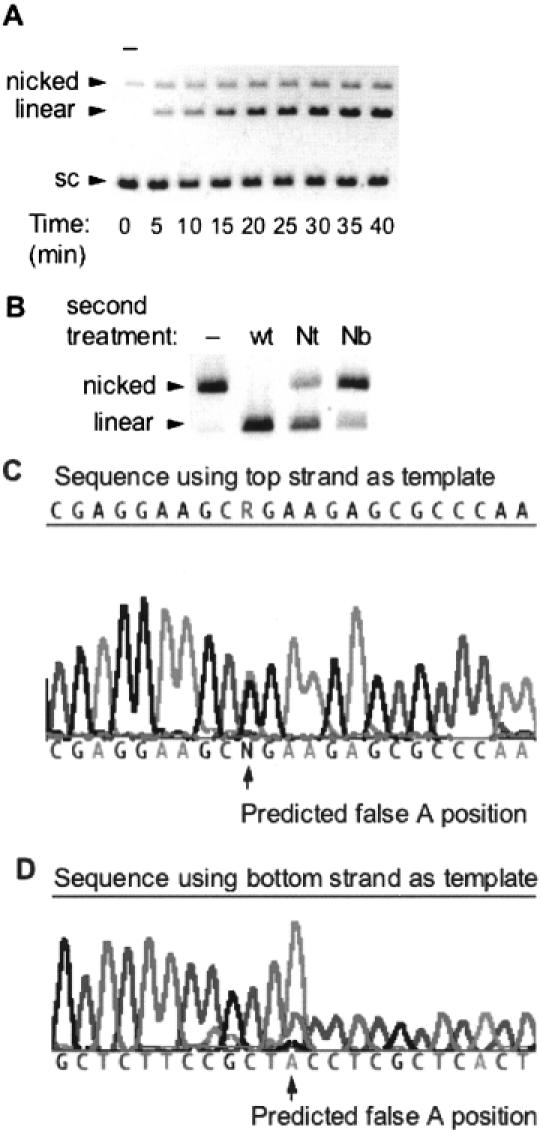
(A) DNA nicking/cleavage characteristics of wt SapI. SapI at a concentration of 1 nM was incubated with 8 nM pUC19 in a 200 μl reaction incubated at 37°C for 40 min. Aliquots of 20 μl were withdrawn every 5 min and added to 5 μl stop solution. Lane 1 is an aliquot taken before enzyme addition (−). (B) Characterization of the nicks created by incubating limiting amounts of wt SapI with pUC19 [as displayed in (A)]. Pre-nicked pUC19 was gel-purified and then subsequently incubated with Nt.SapI-1 (lane 3) or Nb.SapI-1 (lane 4) to determine the strand specificity of the pre-existing nick. Lane 2 is a control digest with excess wt SapI. (C) Sequence of pUC19 nicked by wt SapI. The R indicates a mixed peak of G from circular templates and A from linearized (nicked) top strands. (D) Sequence results using the bottom strand as template. Note the large false A peak (from nicked templates) at the same position as a small T peak produced from circular templates.
