Figure 5.
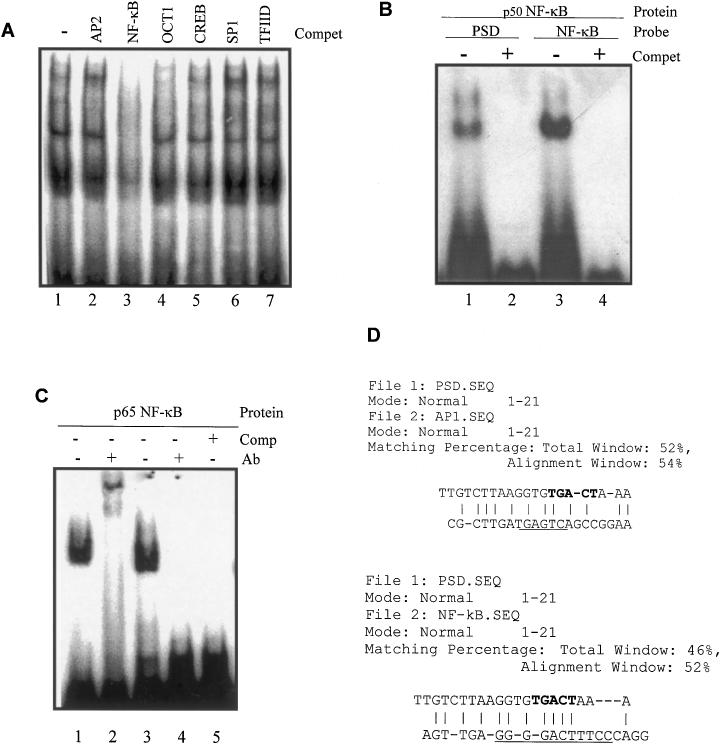
Analysis of transcription factors binding to the PSD oligonucleotide by competitive EMSA. (A) Of the end-labeled PSD nucleotide, 0.03 ng (30 000 c.p.m.) was incubated with 10 μg PC-3 cell nuclear extract proteins in the absence (lane 1) or presence (lanes 2–7) of 100-fold molar excess of different unlabeled consensus oligonucleotides. The 32P-labeled PSD DNA–protein complex formation was conducted in the presence of AP2 (lane 2), NF-κB (lane 3), OCT1 (lane 4), CREB (lane 5), SP1 (lane 6) or TFIID (lane 7) consensus oligonucleotides. (B) Electrophoretic mobility experiments using 32P-labeled PSD and consensus NF-κB oligonucleotides. Labeled oligonucleotides were incubated with 1 × 10−3 gsu [1 gsu = the amount of protein required to gel shift the NF-κB oligo under defined conditions (Promega)] purified p50 NF-κB protein in the absence (−) or presence (+) of 100-fold molar excess of the unlabeled corresponding oligonucleotide. Lanes 1 and 2, end-labeled PSD oligonucleotide incubated with purified p50 NF-κB protein in the absence (−) or presence (+) of 100-fold molar excess of unlabeled PSD oligonucleotide; lanes 3 and 4, end-labeled NF-κB consensus oligonucleotides incubated with purified p50 NF-κB protein in the absence (−) or presence (+) of 100-fold molar excess of unlabeled NF-κB consensus oligonucleotide. (C) Electrophoretic mobility super-shift experiments using labeled PSD and consensus NF-κB oligonucleotides. Labeled oligonucleotides were incubated with the p65 NF-κB protein in the presence or absence of the anti-p65 NF-κB polyclonal antibody. Lanes 1 and 2, end-labeled NF-κB consensus oligonucleotides were incubated with 1 × 10−3 gsu purified p65 NF-κB protein in the absence (−) or presence (+) of the anti-NF-κB Ab; lanes 3 and 4, the end-labeled PSD oligonucleotide was incubated with purified p65 NF-κB protein in the absence (−) or presence (+) of the anti-NF-κB Ab; lane 5, the end-labeled PSD nucleotide was incubated with 1 × 10−3 gsu purified p65 NF-κB protein in the presence of 100-fold molar excess of unlabeled PSD oligonucleotide. (D) Computer alignments of the PSD oligonucleotide sequence with the AP1 or NF-κB consensus sequences in the commercial oligonucleotides were analyzed according to the Needleman–Wunsch algorithm using the DNASIS sequence analysis software (Hitachi). The maximal match was achieved by the insertion of gaps, designated as hyphens. Vertical bars indicate matching regions. The highest score is presented. The consensus AP1 and NF-κB-binding sites are underlined. The putative AP1 binding site in the PSD oligonucleotide was shown in bold.
