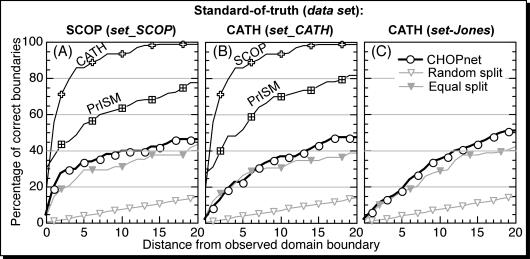
Figure 2.
Accuracy of predicting the precise domain boundary. For all two-domain proteins for which the domain number was predicted correctly, the distance between predicted and observed domain boundaries was measured, and the accuracy of the prediction was calculated for thresholds ranging from 0–20 residues and compared with that of two trivial predictions: ‘random split’ and ‘equal split’. The left panel (A) gives the results for using SCOP assignments as standard-of-truth. The lines for CATH and PrISM mark the agreement between SCOP and these two methods that are based on known 3D structures. The two panels on the right (B and C) are based on using CATH assignments as standard-of-truth. Panels (A and B) were based on the cross-validation section common between CATH and SCOP (set_SC), while (C) was based on the test set used to evaluate DomSSEA (47).