Abstract
The detailed chemical information contained in the vibrational spectrum of a cryogenically cooled analyte would, in principle, make infrared (IR) ion spectroscopy a gold standard technique for molecular identification in mass spectrometry. Despite this immense potential, there are considerable challenges in both instrumentation and methodology to overcome before the technique is analytically useful. Here, we discuss the promise of IR ion spectroscopy for small molecule analysis in the context of metabolite identification. Experimental strategies to address sensitivity constraints, poor overall duty cycle, and speed of the experiment are intimately tied to the development of a mass-selective cryogenic trap. Therefore, the most likely avenues for success, in the authors? opinion, are presented here, alongside alternative approaches and some thoughts on data interpretation.
Keywords: vibrational spectroscopy, action spectroscopy, cryogenic, metabolites, ion trap
Introduction
Many mass spectrometry technique developments have aimed to increase the amount of structural information obtained through these measurements. In ion spectroscopy, the structure of an ion is probed via photons from an intense light source, usually a laser. The absorption of light is most readily verified by a so-called ‘action’ or ‘consequence’ spectroscopy scheme, in which the absorbed photon increases the ion’s internal energy, thereby (possibly) leading to cleavage of a bond and a change in its mass-to-charge ratio (m/z). The yield of this photodissociation process is dependent on properties of the light source, such as the photon wavelength/energy and the spectral brightness, but is also importantly reliant on the molecular properties of the ion, namely its electronic and vibrational absorption characteristics. As has been demonstrated in countless publications since the late 1970’s, indirect measurements of the infrared (IR) and/or ultraviolet-visible (UV-Vis) spectra of ions can be obtained via wavelength-dependent laser-induced photodissociation.
Progress in infrared ion spectroscopy was held back for a long time by the limited tunability of light sources. In the past decade and a half, the field has enjoyed a considerable resurgence, initially brought about by the advent of widely tunable free electron lasers (FELs), housed at user facilities (e.g. FELIX, CLIO) [1, 2], which have allowed for the recording of IR spectra in the chemically important 500-2000 cm−1 so-called ‘fingerprint’ region. More recently, benchtop optical parametric oscillators/amplifiers (OPO/As) have become much more routinely operated, and have extended the capability of recording IR photodissociation spectra (in the e.g. 2150-4400 cm−1 region) to individual investigator laboratories at an affordable cost (i.e., <$100k) [3-5]. By using appropriate non-linear crystals (e.g. AgGaSe2) [6], output can also be generated in the important 550-2000 cm−1 region, in principle spanning the entire range that is usually probed by FTIR (Fourier transform infrared) spectroscopy. Given these developments, and a recent review covering this topic [7], a detailed discussion on IR light sources will not be the subject of this paper, but the focus will rather be on mass spectrometry developments.
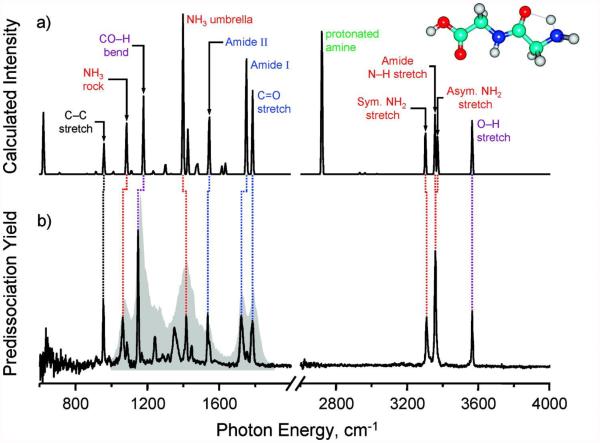
Despite these advances, why then has ion spectroscopy still not emerged as a powerful tool in (bio)analytical mass spectrometry? A key impediment is the relatively poor resolution of the most easily implemented IR ‘action’ spectroscopy method, infrared multiple photon dissociation (IRMPD) of room-temperature ions [8-10], which generally gives rise to broad IR bands with full-width half-maximum (FWHM) in the range of 30-50 cm−1. Enhanced resolution (i.e., FWHM ≤6 cm−1), and thus higher molecular specificity, can be achieved in cryogenic experiments, where the ion structures are probed at temperatures close to absolute zero (i.e., ~4-30K) [11, 12]. At these low temperatures, spectral congestion is noticeably reduced, due to the elimination of hot bands, and a narrower rotational distribution. As an example, the experimental IR spectra for protonated diglycine at room temperature (using IRMPD spectroscopy) and cryogenic temperature (using IR predissociation spectroscopy) are contrasted in Figure 1 [13], together with the computed IR spectrum of the lowest-energy conformer. The cryogenic measurements clearly exhibit a much closer agreement with theory, and thus a more confident structural assignment. As a word of caution, however, a number of conformers may be generated in the cooling process, especially in the case of larger molecules, meaning that the observed IR spectrum is due to a co-addition of all of these conformers.
Figure 1.
Comparison of computed IR absorption spectrum of protonated diglycine (top), with experimental IRMPD (gray shaded) and cryogenic (black line) IR spectra of the same molecule (bottom). Adapted with permission from Kamrath, M.Z., Garand, E., Jordan, P.A., Leavitt, C.M., Wolk, A.B., Stipdonk, M.J.V., et al.: Vibrational Characterization of Simple Peptides Using Cryogenic Infrared Photodissociation of H2-Tagged, Mass-Selected Ions. J Am Chem Soc. 133, 6440–6448 (2011). Copyright 2011 American Chemical Society.
Despite the more detailed structural information that is obtainable in these experiments, cryogenic measurements also require custom instrumentation that is not readily available commercially. In fact, even current, cutting-edge cryogenic instrumentation (for ion spectroscopy) displays considerable shortcomings in terms of turning the technique into a useful analytical tool. For one, less emphasis has so far been put on maximizing sensitivity in cryogenic instruments, in order to make the technique viable for ‘real’ biological samples. Moreover, ‘action’ spectroscopy approaches generally suffer from a poor duty cycle, as a single precursor ion is monitored at a single laser frequency at any one time. Finally, the chemical identification of truly unknown analytes, based on their infrared spectra, is a non-trivial problem, as the number of putative candidate structures (based on mass) is extremely large [14].
This critical insight article aims to offer possible solutions in mass spectrometry instrumentation and methodology to overcome these challenges. The detailed structural insights that infrared ion spectroscopy of cryogenically cooled ions offers, and the potentially revolutionary implications of these developments for small molecules analysis (such as in metabolomics), suggest that these efforts are more than worthwhile. It should also be noted that there are currently a number of exciting strategies involving spectroscopy that are on-going with the aim of increasing the structural information on biological ions. These include, among others, the coupling of ion mobility with cryogenic spectroscopy [15, 16], as well as using the distinct UV photodissociation fingerprints of cryogenically cooled ions [17] (as a function of wavelength) for purposes of identification.
The Promise of a New Technique for an Old Problem
An infrared spectrum of an ion contains a wealth of chemically relevant information, such as: the presence or absence of various chemical groups, the symmetry of the complex, the site of the charge attachment, the strength of hydrogen bonding interactions, and the secondary structure of peptides/proteins (to name a few). Spectral congestion generally increases with molecular size, due to a larger number of oscillators, but also due to the possible presence of multiple conformers. This is further compounded by a larger number of isomers for larger molecular weights, meaning that more analytes need to be differentiated at higher masses. These trends suggest that there is an upper-mass limit, above which unambiguous molecular identification via IR spectroscopy is difficult, if not impossible. In order to avoid this problem, we aim to apply the technique primarily to small-molecule analysis.
Metabolomics involves profiling small molecular weight analytes found in biological systems. The high complexity of metabolite samples poses a formidable challenge for the structural identification of unknowns [18, 19]. Current methodologies involving liquid chromatography (LC) separation, followed by MS/MS fragmentation merely allow a small proportion of metabolites to be identified [20, 21]. The main bottleneck lies in the limited number of standards for which reference MS/MS spectra are available, compounded by the instability of many metabolites.
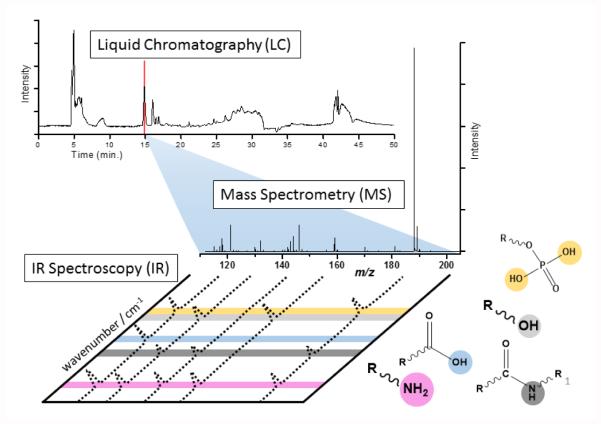
The application of IR ion spectroscopy to metabolite identification, as shown in Figure 2, introduces an additional dimension of information. Some of this information can be relatively straightforward to interpret, such as the presence of various diagnostic bands, and thus the presence (or absence) of various chemical groups. For instance, the band positions of a number of moieties have been found to be relatively reproducible in peptides, as summarized in Table 1.
Figure 2.
Schematic view of LC-MS analysis in metabolomics, followed by IR ion spectroscopy analysis of selected metabolite features. Diagnostic vibrational modes are depicted by color coding.
Table 1.
Summary of diagnostic band positions for various moieties in peptide systems, based on previous studies [13, 16, 22-30].
| Chemical moiety and vibrational mode | Band positions |
|---|---|
| Amide NH bend (Amide II) | 1475-1525 |
| Amide CO stretch (Amide I) | 1675-1725 |
| Carboxylic acid CO stretch | 1725-1775 |
| Indole NH stretch | 3480-3520 |
| Carboxylic acid OH stretch | 3540-3600 |
| Sulfate OH stretch | 3580-3600 |
| Alcohol OH stretch | 3600-3675 |
| Phosphate OH stretch | 3650-3700 |
Particularly the so-called fingerprint region (i.e., 500-2000 cm−1) can serve as a strong identifier for molecular identification, as is well known from Fourier Transform IR (FTIR) spectroscopy. The higher spectral resolution in cryogenic spectroscopy is important to increase the molecular specificity in these measurements. Another important characteristic of IR spectroscopy is that molecules with similar chemical structures exhibit similarities in their spectral patterns. This is likely key in the identification of unknown metabolites.
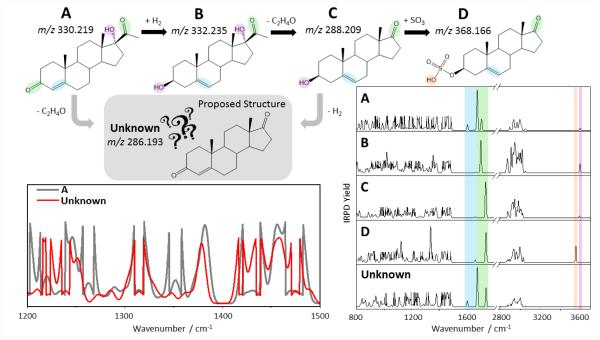
Figure 3 exemplifies the power of the combined information from mass spectrometry and IR spectroscopy to make structural assignments for unknown metabolites. Known metabolites from the androgen/estrogen metabolic cycle, denoted here as A-D, are employed for illustrative purposes. The various known metabolites in this cycle are naturally closely related chemically, as each metabolic step (merely) changes some functional groups, but leaves the main backbone structure intact. Let’s assume that an unknown metabolic feature were measured at an accurate m/z 286.193. Based on mass alone, there are in principle a very large number of putative structures that would need to be considered, and it would not be obvious that this molecule is related to the known molecules A-D. If, however, the IR spectrum of the unknown were compared to reference IR spectra for A-D, one would expect a considerable degree of spectral similarity. This is illustrated in more detail for the 1200-1500 cm−1 region, where the unknown is contrasted to the known structure A, showing a number of bands that are present in both spectra. In general, one would expect that metabolites within the same chemical family would exhibit considerable spectral similarities, especially in the fingerprint region, which is largely composed of backbone deformation and bending modes. This characteristic match would allow a general classification of the unknown molecule within a particular chemical family, provided that suitable reference spectra (of known analytes) are available. In addition, the accurate mass measurement would enable an assignment of the elemental composition, in effect guiding the process of proposing putative structures for the unknown within a given family. More chemically diagnostic modes, such as carbonyl CO stretches, C=C stretches, OH stretches, etc. (denoted by color coding), could then verify which chemical moieties are present/absent, thus further reducing the number of putative structures that need to be considered. An ultimate identification of the unknown is achievable by matching the IR spectrum of the unknown to that of a synthetically made reference compound. Given the high resolution in the cryogenic experiments, an exact overlap could serve as a convincing identifier, even if some detailed questions, such as those related to stereochemistry, may remain unanswered.
Figure 3.
Illustrative metabolic pathway with known metabolites A-D, and proposed structure for unknown metabolite (top). Comparison of experimental IR spectrum of unknown with reference experimental IR spectra for known metabolites (right), depicting diagnostic vibrations via color coding. Zoom-in of 1200-1500 cm−1 region overlay for known A and unknown (left bottom).
(This illustrative figure was made using the androgen/estrogen metabolic pathway as an example, incorporating the known metabolites 17-hydroxyprogesterone (A), 17α-hydroxypregnenolone (B), dehydroepiandrosterone (C), Dehydroepiandrosterone sulfate (D), and androstenedione (unknown). The IR spectra for these molecules were obtained in the Gaussian 03 quantum chemical package, using the B3LYP/6-31G* level of theory for geometry optimization and harmonic frequency calculations. Note that in the region from 800-1500 cm−1, the intensities were magnified by a factor between 3-10 for ease of visual representation. The frequencies were scaled by a uniform scaling factor of 0.961, except for OH stretches, where a scaling factor of 0.976 was employed. The stick spectra were then convoluted using a 5 cm−1 full-width half-maximum (FWHM) Gaussian function to make them appear more similar to experimental data.)
A similar argument for the usefulness of IR spectroscopy can be made in the area of testing for banned substances. Traditional methodologies for drug testing, based on chromatographic retention times, accurate mass and fragmentation patterns, backed up by optimized sample work-up procedures, are highly selective and proficient for determining the concentrations of drugs with known molecular formulae. ‘Designer drugs’ are engineered in such a way as to evade detection by these testing methods [31, 32], by shifting retention times and the masses of analytes, and thus pose a problem for analytical science. Nonetheless, a potential Achilles heel of designer drugs is the fact that they must retain certain chemical properties, namely their relevant chemical functional groups and/or structure, in order to avoid sacrificing their effectiveness. It stands to reason that IR spectroscopy should be well placed to bring to the fore those molecules that are most chemically similar to known banned substances, and thus act as a powerful method for drug analysis.
While there are many other fruitful avenues for IR ion spectroscopy, the usefulness of the technique is constrained by molecular size – the large number of oscillators in a protein, for instance, can render a detailed structural interpretation challenging. Still, this does not mean that answers to more selective questions cannot be obtained. For instance, the presence of functional groups with distinct vibrational features, such as oxazolone rings [33, 34], and phosphates [26, 35, 36] have been verified in larger peptides. Further, IR ion spectroscopy is powerful for determining the secondary structure motifs of peptides (even up to 12 amino acid residues) [37]. The driving forces for advancing a particular technology can be found in both the strengths of the novel technology as well as the failings of traditional approaches. In this light, metabolites and banned substances rank among the most manifest examples, where further analytical methodology developments are required.
General Challenges
The key challenge in applying IR ion spectroscopy to biological research lies in sample consumption. As a mass spectrometric technique, the inherent sensitivity of ion spectroscopy is high, certainly in contrast to the structural competitor technique nuclear magnetic resonance (NMR). On the other hand, as an IR ion spectrum consists of photodissociation mass spectra at several, perhaps hundreds of discrete laser frequency steps, this naturally reduces the sensitivity of the methodology (when compared to the acquisition of a single mass spectrum). The problem boils down to the extremely low duty cycle of IR ion spectroscopy, as at any one time, a single precursor is probed at a single laser frequency. One way to overcome this problem lies in multiplexing the experiment, such that the IR spectra of multiple analytes are measured in parallel. A related problem lies in the speed with which IR spectra can be recorded, which also affects sample consumption.
Considerations on Experimental Methodologies and Instrumentation
The challenges above raise a number of questions. Which IR “action” spectroscopy scheme is most useful within a bioanalytical methodology? Which type of a mass spectrometer would be best suited for these measurements? What trade-offs are there in terms of maximizing sensitivity, speed, and spectral resolution?
A key requirement is that the spectroscopic technique is performed at higher resolution, necessitating cryogenically cooled ions. Among the various cryogenic ion IR spectroscopy schemes, the messenger spectroscopy method, also known as predissociation spectroscopy [36], is, in our opinion, best positioned for bioanalytical applications. Briefly, in messenger spectroscopy, atoms or molecules are attached to analyte ions via weak van der Waals interactions at cryogenic temperatures; the tag is then detached upon resonant absorption of a photon by the analyte ion.
The advantages of messenger spectroscopy are multifold. First and foremost, the methodology is applicable to any molecule, as IR modes of the molecule are probed. In contrast, for instance the IR-UV double resonance scheme [38, 39] or IR photofragment gain spectroscopy [40] requires the presence of a UV chromophore (with a suitably long lifetime of the excited state) [41], thereby constraining the types of molecules that can be probed. Another key advantage of the tagging scheme ensues from the known mass of the tagging gas, which, barring any mass interferences, allows for simultaneous tagging of multiple analyte molecules at different m/z. These tagged analyte ions can then be subjected to laser irradiation, and the individual photodissociation behavior of several analytes can be investigated in the same experiment. This sets the stage for a multiplexed IR ‘action’ spectroscopy scheme. A final advantage of messenger spectroscopy is that the technique does not rely on ultimately cold temperatures (i.e., close to 4K), at least if slightly stronger-binding tags (e.g. H2, N2, Ne, Ar) are employed. In practice, this means that ion tagging can be achieved in more conventional mass spectrometric traps, such as 3D ion traps, as demonstrated in a number of studies [42, 43]. Crucially, the latter traps exhibit powerful mass-selective capabilities, allowing for mass manipulation and detection of ions inside the cryogenic trap. On a sidenote, there has been a general trend in cryogenic spectroscopy to lower-order traps, such as octopoles [44] and even (wire-type) quadrupoles [45], which nonetheless achieve low temperatures, mainly for the purpose of enhancing the overlap of the ion cloud with the laser beams; even if those particular traps do not display mass-selective capabilities (yet). There are also some disadvantages of messenger spectroscopy vis-á-vis IR-UV double resonance spectroscopy; the tag can cause a distortion in the (inherent) IR spectrum of the ion, and it is not as convenient to select a particular conformer, even if IR-IR approaches [46] are capable of “burning” away particular conformers. In terms of using messenger spectroscopy as an analytical tool, where structural identification can be achieved via IR spectral matching to known library compounds, the spectral distortion and presence of multiple conformers may not represent an impediment however, provided that the van der Waals tag is the same in both measurements, meaning that the distortion is also identical.
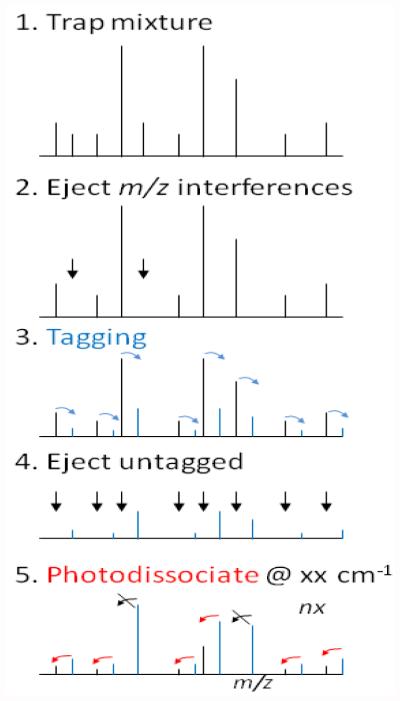
Figure 4 illustrates an idealized scheme for multiplexed IR spectroscopy for a mixture of analytes, using the tagging approach. After trapping (1), mass interferences are removed (2), to prevent mass overlaps with predicted masses of tagged ions. A gas pulse containing the tagging agent will then cool the ions and convert some ion population into tagged ions (3). The remaining portion of untagged ions are mass ejected (4), followed by laser irradiation at a particular frequency (5). Some ions will dissociate at this frequency (red arrows), whereas others do not, reflecting their IR absorbance characteristics. Note that the ejection of the precursor in step (4) is essential for a background-free detection of the photodissociation, thereby maximizing the sensitivity of measurement. This whole procedure is repeated at different laser frequency steps to record the IR spectra of multiple ions in a multiplexed approach. While steps 3-5 would have to be carried out inside a cryogenic trap, steps 1-2 could more easily be performed in an upstream trap, as the ions only need to be cold for tagging purposes. It should be noted that this scheme has so far not been put into practice due to the challenges of mass-manipulating ions in a cryogenic trap, most notably the extreme fragility of tagged ions. These challenges will be discussed in more detail below.
Figure 4.
Multiplexing scheme in messenger spectroscopy
What is the ideal tag in these experiments?
From a spectroscopic point of view, the ideal tag is one that causes the least distortion in the inherent IR spectrum of the analyte ion. In other words, the tag that is the least polarizable and therefore has the lowest binding energy. From that point of view, helium would be the ideal choice. From a mass spectrometry point of view, however, tagging with helium is challenging. Helium will only tag at extremely low temperatures (i.e., <10K), and has so far not been implemented in cryogenic traps with mass-selective capabilities. Moreover, the mass shift by helium (4 Da), or for that matter hydrogen (2 Da) or deuterium (4 Da), are low, meaning that mass-ejecting untagged ions (in step 4 of Figure 4) is likely to result in a non-resonant activation (i.e., acceleration) of the tagged ions. As the tagged ions are extremely fragile, any collisional activation will lead to loss of the tag, and thus a non-radiative decay of the target ion population. In a best-case scenario, this undesirable process will add significant noise to the IR spectra by spoiling the background-free detection scheme; in a worst-case scenario, the tagged ion population will be completely depleted, precluding the measurement of the IR messenger spectrum altogether!
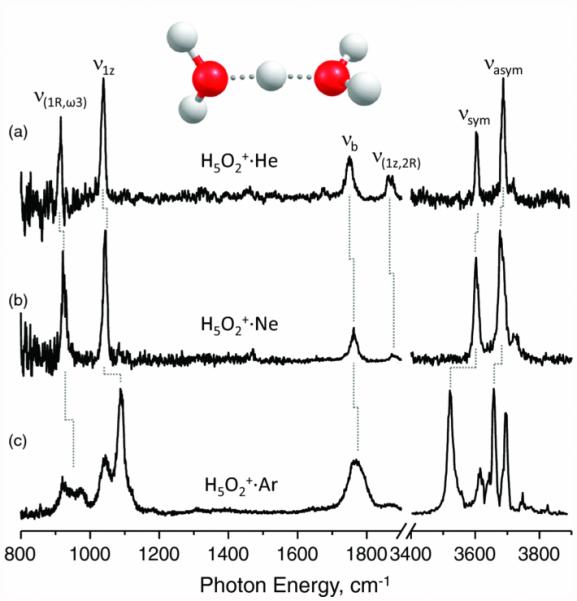
The use of higher-mass tags is advisable in terms of simplifying mass ejection of the untagged ions remaining, while still minimizing activation of the tagged ions. Nitrogen (28 Da) and argon (40 Da) are natural choices, given their low cost, and previous use in messenger spectroscopy. Neon (20 Da) is also a viable candidate, as it exhibits fewer distortions of the IR spectrum than argon, while condensing at intermediate temperatures between helium and argon. The effect of the tag on the IR messenger spectrum is illustrated in Figure 5 for the example of the proton-bound dimer of water, H5O2+. The Ne-tag spectrum appears similar to the one using He-tags, whereas the Ar-tag causes a number of spectral distortions. On a sidenote, the Ne- and Ar- tagging in Figure 4 was achieved in a supersonic expansion [47]. While it is clear that the tag can affect the IR spectrum of an analyte, it should be noted that proton-bound complexes are also extreme examples in IR spectroscopy. The floppy nature of these molecular systems, and the highly anharmonic, large-amplitude motions of the shared proton allows for coupling interactions that are vastly more pronounced than for other (covalently bound) molecules.
Figure 5.
Comparison of IR messenger spectra of proton-bound water dimer H5O2+ tagged with He, Ne and Ar. Reprinted with permission from Johnson, C.J., Wolk, A.B., Fournier, J.A., Sullivan, E.N., Weddle, G.H., Johnson, M.A.: Communication: He-Tagged Vibrational Spectra of the SarGlyH+ and H+(H2O)2,3 Ions: Quantifying Tag Effects in Cryogenic Ion Vibrational Predissocation (CIVP) Spectroscopy. J Chem Phys. 140, 22 (2014). Copyright 2014, AIP Publishing LLC.
The ideal tag for bioanalytical applications also depends on other parameters that affect the overall ease of measuring these spectra, such as the tagging efficiency. The conversion rate of untagged ions into tagged ions is governed primarily by the temperature of the ions and number density of the tag molecules. A complicating factor in these experiments is that the tag molecules can ‘freeze out’ on the electrodes, causing a distortion in the electric fields, and thus malfunction of the ion trap in the longer run. For this reason, the tag molecules are often added as a small component in a helium gas mix. The role of helium being to collisionally thermalize the ions, which requires many collisions with the trap electrodes and the ions. Helium is a natural choice for this purpose, as it will not ‘freeze out’ on the electrodes. The optimum tag gas concentration will depend on the design of the trap and the trade-offs of maximizing the yield of singly-tagged ions, while still allowing the ion trap to be operated for a number of hours. To put the discussions above into context, so far, He [48, 49], H2 [4, 16], D2 [46], Ne [48], and N2 [48] have already been shown to be successfully tagged onto ions in cryogenic traps, but this feat remains to be implemented for higher-temperature melting point gases, such as for instance Ar.
What is the ideal cryogenic trap?
Conservatively, the yield of singly-tagged ions can be as high as a few percent, meaning a net loss of >90% of ion signal upon going from untagged to tagged ions. The ideal mass spectrometer for messenger spectroscopy, carried out on a number of analytes simultaneously, thus involves a mass-selective cryogenic trap with a high dynamic range of detection and powerful MS/MS capabilities. Table 2 lists some of the ion traps that can be considered for this purpose, along with key characteristics for maximizing sensitivity. We will discuss the merits for each of these possibilities in turn. Note that due to the constraints in these experiments, hybrid designs, involving additional upstream traps, are likely even more effective in terms of facilitating these measurements.
Table 2.
Comparison of ion traps in terms of sensitivity constraints. + denotes enhanced performance compared to −.
| Characteristics | 3D ion trap | 2D linear ion trap | FTICR |
|---|---|---|---|
| Trapping efficiency | − | + | + |
| Trapping capacity | − | + | + |
| Detection limit | + | + | − |
The experiments by Johnson and co-workers have been carried out on a cryogenically cooled 3D Paul ion trap coupled to a time-of-flight (TOF) analyzer, and thus it is clear that ion tagging can be successfully implemented in these types of traps [4].
A linear ion trap (LIT) (with mass selection capabilities), also known as a 2D ion trap, is generally considered among the most powerful tools for MSn experiments, given its high trapping efficiency (i.e., close to 100% compared to 1-10% for 3D ion traps) and capacity (i.e., higher by an order of magnitude than 3D ion trap), extensive mass isolation capabilities, and low detection limits [50-53]. In a practical sense, the large number of initially trapped analyte ions make these traps tolerant to ion losses as a result of low conversion rates. Despite their considerable potential, mass-selective LITs have so far not been implemented for cryogenic IR spectroscopy applications.
Another possibility as a cryogenic ion trap is a Fourier transform ion cyclotron resonance (FTICR) trap, which has the advantage of high vacuum operation and excellent mass manipulation characteristics. FTICR traps have already been demonstrated in cryogenic experiments, at least down to 133K [54, 55]. A disadvantage of the induced current detection scheme for FTICR is the lower sensitivity vis-á-vis electron multiplier detection (by about two orders of magnitude) [56].
A complicating factor in these experiments is the need for collisional cooling of the ions, in order to form the tagged complex. At cryogenic temperatures, the neutral density of the pulsed gas remains high for extended periods (tens to hundreds of ms) [57], and this poses a potential problem for precluding dissociative collisions of the fragile messenger ions. In the experiments by Johnson et al., the tagged ions were not irradiated inside the 3D ion trap, but rather in the TOF analyzer, using the TOF for mass separation and thus mass selection of the tagged species [13]. That approach minimizes problems with dissociation of the tagged ions due to collisions with background gas, as the background pressure in the TOF chamber is considerably lower than in the 3D ion trap. In terms of implementing IR action spectroscopy inside of the cryogenic trap, there is a clear trade-off between maximizing ion cooling/tagging on the one hand, while minimizing fragmentation of the tagged ions on the other. Another option is to utilize two cryogenic ion traps, one for cooling and tagging, operated at a higher pressure, and one for mass isolation, photodissociation, and mass detection, operated at a lower pressure. At this point, it remains to be seen which instrumentation offers the most facile implementation, while remaining a cost effective option. From a point of view of maximizing sensitivity, it is our opinion that cryogenic LITs will play a prominent role to address this challenge, and in fact dual LIT designs are already commercially available, and fit well into this scheme [58].
At what speed can IR spectra be recorded?
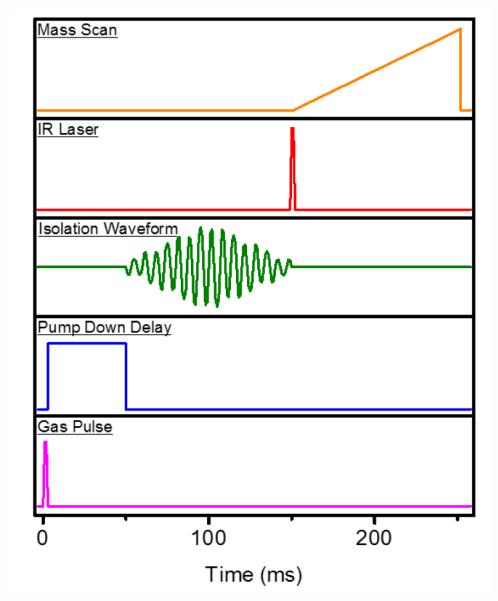
The answer to this question is intimately tied to the time required for taking a measurement at a single laser frequency. In the experimental sequence in Figure 6, rough estimates for various events are indicated. For reference, the reader may compare these timings to alternative cryogenic spectroscopy approaches and instrumentation [17, 59]. A likely bottleneck in this sequence is the pump down delay, which must at least be on the order of several tens of ms, but may have to exceed 100 ms, in order to achieve a lower background pressure during ion manipulation (i.e., mass isolation and mass scan). Assuming a best-case scenario of ~4 Hz, at which mass spectra can be recorded, and the need for a single mass spectrum at a laser step size of 1 cm−1, to be recorded in duplicate, the total scan time for a mid-IR spectrum from 800-1800 cm−1 would be on the order of 500 s, or just under 9 minutes. For comparison, Kopysov et al. reported a measurement time of 150 ms per wavelength point in their UV photodissociation experiments [17].
Figure 6.
Time sequence of mass spectrometry experiment for IR ‘action’ spectroscopy inside cryogenic trap (from bottom to top).
For what types of samples would these measurements be useful?
Given the rather lengthy scan times, it is expected that these experiments will not be compatible with LC elution profiles for the foreseeable future. Instead, it makes more sense that high-value LC fractions, containing potential biomarkers that are yet unidentified using current methodologies, are carried out offline. Ideally, the complementary mass measurements with conventional approaches are carried out at high mass resolution and high mass accuracy, in order to enhance molecular specificity from the combined mass and IR information, but also avoid the presence of multiple isobaric species under one unit mass.
Summary
While there have been substantial advances in measuring high resolution IR action spectra of biomolecular ions over the past decade, the primary focus of those experiments has been on obtaining a fundamental understanding of gas-phase structure, intimately tied in with theoretical predictions of structure, and thus a benchmarking of theory. This critical insight article has attempted to give a perspective for bioanalytical applications of similar technologies and methodologies. It is proposed to implement IR messenger spectroscopy in the context of small molecule identification, illustrated primarily for the example of metabolite identification. The tagging approach in messenger spectroscopy is, in principle, suited to recording the IR spectra of multiple analytes in a multiplexed fashion. Such a scheme would most easily be implemented in a mass-selective cryogenic trap, where all ions are tagged and spectroscopically probed in parallel. A cryogenic linear ion trap with mass selection capabilities is expected to be the ultimate tool for performing ion spectroscopy experiments on mass-separated ions, due to its high sensitivity, and powerful MS/MS capabilities. The choice of the ideal mass tag remains to be investigated, in the context of providing high-quality, high-resolution IR spectra, as well as ease of implementation in a cryogenic trap.
As challenges specific to instrumentation are overcome, we anticipate that the first demonstration of IR ion spectroscopy as a bioanalytical tool will take place in the next few years. This will hopefully spur commercial manufacturers to produce mass spectrometry instrumentation that is more suitable for ion spectroscopy measurements, notably mass-selective cryogenic ion traps.
Acknowledgments
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM110077. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Oepts D, van der Meer AFG, van Amersfoort PW. The free-electron-laser user facility FELIX. Infrared Physics & Technology. 1995;36:297–308. [Google Scholar]
- 2.Lemaire J, Boissel P, Heninger M, Mauclaire G, Bellec G, Mestdagh H, et al. Gas Phase Infrared Spectroscopy of Selectively Prepared Ions. Phys. Rev. Lett. 2002;89:273002. doi: 10.1103/PhysRevLett.89.273002. [DOI] [PubMed] [Google Scholar]
- 3.York RL, Holinga GJ, Guyer DR, McCrea KR, Ward RS, Somorjai GA. A New Optical Parametric Amplifier Based on Lithium Thioindate Used for Sum Frequency Generation Vibrational Spectroscopic Studies of the Amide I Mode of an Interfacial Model Peptide. Applied Spectroscopy. 2008;62:937–940. doi: 10.1366/000370208785793227. [DOI] [PubMed] [Google Scholar]
- 4.Kamrath MZ, Relph RA, Guasco TL, Leavitt CM, Johnson MA. Vibrational predissociation spectroscopy of the H2-tagged mono-and dicarboxylate anions of dodecanedioic acid. International Journal of Mass Spectrometry. 2011;300:91–98. [Google Scholar]
- 5.Gerhards M. High energy and narrow bandwidth mid IR nanosecond laser system. Optics Communications. 2004;241:493–497. [Google Scholar]
- 6.Abedin KS, Haidar S, Konno Y, Takyu C, Ito H. Difference frequency generation of 5–18 μm in a AgGaSe2 crystal. Applied Optics. 1998;37:1642–1646. doi: 10.1364/ao.37.001642. [DOI] [PubMed] [Google Scholar]
- 7.Stedwell CN, Galindo JF, Roitberg AE, Polfer NC. Structures of Biomolecular Ions in the Gas Phase Probed by Infrared Light Sources. Annual Review of Analytical Chemistry. 2013;6:267–285. doi: 10.1146/annurev-anchem-062012-092700. [DOI] [PubMed] [Google Scholar]
- 8.Oomensa J, Sartakova BG, Meijerc G, Helden G.v. Gas-phase infrared multiple photon dissociation spectroscopy of mass-selected molecular ions. International Journal of Mass Spectrometry. 2006;254:1–19. [Google Scholar]
- 9.Polfer NC. Infrared Multiple Photon Dissociation Spectroscopy of Trapped Ions. Chemical Society Reviews. 2011;40:2211–2221. doi: 10.1039/c0cs00171f. [DOI] [PubMed] [Google Scholar]
- 10.Fridgen TD. Infrared Consequence Spectroscopy Of Gaseous Protonated And Metal Ion Cationized Complexes. Mass Spectrometry Reviews. 2009;28:586–607. doi: 10.1002/mas.20224. [DOI] [PubMed] [Google Scholar]
- 11.Rizzo TR, Stearns JA, Boyarkin OV. Spectroscopic studies of cold, gas-phase biomolecular ions. International Reviews in Physical Chemistry. 2009;28:481–515. [Google Scholar]
- 12.Garand E, Kamrath MZ, Jordan PA, Wolk AB, Leavitt CM, McCoy AB, et al. Determination of Noncovalent Docking by Infrared Spectroscopy of Cold Gas-phase Complexes. Science. 2012;335:694–698. doi: 10.1126/science.1214948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamrath MZ, Garand E, Jordan PA, Leavitt CM, Wolk AB, Stipdonk MJV, et al. Vibrational Characterization of Simple Peptides Using Cryogenic Infrared Photodissociation of H2-Tagged, Mass-Selected Ions. Journal of the American Chemical Society. 2011;133:6440–6448. doi: 10.1021/ja200849g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blum LC, Reymond J-L. 970 Million Druglike Small Molecules for Virtual Screening in the Chemical Universe Database GDB-13. Journal of American Chemical Society. 2009;131:8732–8733. doi: 10.1021/ja902302h. [DOI] [PubMed] [Google Scholar]
- 15.Papadopoulos G, Svendsen A, Boyarkin OV, Rizzo TR. Conformational Distribution of Bradykinin [bk + 2 H]2+ Revealed by Cold Ion Spectroscopy Coupled with FAIMS. Journal of the American Society for Mass Spectrometry. 2012;23:1173–1181. doi: 10.1007/s13361-012-0384-0. [DOI] [PubMed] [Google Scholar]
- 16.Masson A, Kamrath MZ, Perez MAS, Glover MS, Rothlisberger U, Clemmer DE, et al. Infrared Spectroscopy of Mobility-Selected H+-Gly-Pro-Gly-Gly (GPGG) Journal of the American Society for Mass Spectrometry. 2015;26:1444–1454. doi: 10.1007/s13361-015-1172-4. [DOI] [PubMed] [Google Scholar]
- 17.Kopysov V, Makarov A, Boyarkin OV. Colors for Molecular Masses: Fusion of Spectroscopy and Mass Spectrometry for Identification of Biomolecules. Analytical Chemistry. 2015;87:4607–4611. doi: 10.1021/acs.analchem.5b00822. [DOI] [PubMed] [Google Scholar]
- 18.Ryan D, Robards K. Metabolomics: The Greatest Omics of Them All? Analytical Chemistry. 2006;78:7954–7958. doi: 10.1021/ac0614341. [DOI] [PubMed] [Google Scholar]
- 19.Kell DB. Metabolomics and Systems Biology: Making Sense of the Soup. Current Opinion in Microbiology. 2004;7:296–307. doi: 10.1016/j.mib.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Liu S, Hu Y, Lia P, Wan J-B. Current state of the art of mass spectrometry based metabolomics studies – a review focusing on wide coverage, high throughput and easy identification. RSC Advances. 2015;5:78728–78737. [Google Scholar]
- 21.Halket JM, Waterman D, Przyborowska AM, Patel RKP, Fraser PD, Bramley PM. Chemical derivatization and mass spectral libraries in metabolic profiling by GC/MS and LC/MS/MS. Journal of Experimental Botany. 2005;56:219–243. doi: 10.1093/jxb/eri069. [DOI] [PubMed] [Google Scholar]
- 22.Patrick AL, Stedwell CN, Polfer NC. Differentiating Sulfopeptide and Phosphopeptide Ions via Resonant Infrared Photodissociation. Analytical Chemistry. 2014;86:5547–5552. doi: 10.1021/ac500992f. [DOI] [PubMed] [Google Scholar]
- 23.Schwing K, Reyheller C, Schaly A, Kubik S, Gerhards M. Structural Analysis of an Isolated Cyclic Tetrapeptide and its Monohydrate by Combined IR/UV Spectroscopy. ChemPhysChem. 2011;12:1981–1988. doi: 10.1002/cphc.201001055. [DOI] [PubMed] [Google Scholar]
- 24.Wang D, Gulyuz K, Stedwell CN, Yu L, Polfer NC. Effect of phenol and acidic side chains on the protonation sites of b2 ions confirmed by IRMPD spectroscopy. International Journal of Mass Spectrometry. 2012;330-332:144–151. [Google Scholar]
- 25.Dunbar RC, Steill JD, Polfer NC, Oomens J. Gas-phase infrared spectroscopy of the protonated dipeptides H+PheAla and H+AlaPhe compared to condensed-phase results. International Journal of Mass Spectrometry. 2009;283:77–84. [Google Scholar]
- 26.Correia CF, Balaj PO, Scuderi D, Maitre P, Ohanessian G. Vibrational Signatures of Protonated, Phosphorylated Amino Acids in the Gas Phase. Journal of the American Chemical Society. 2008;130:3359–3370. doi: 10.1021/ja073868z. [DOI] [PubMed] [Google Scholar]
- 27.Gerhards M, Unterberg C, Gerlach A. Structure of a b-sheet model system in the gas phase: Analysis of the C=O stretching vibrations. Physical Chemistry Chemical Physics. 2002;4:5563–5565. doi: 10.1039/b600154h. [DOI] [PubMed] [Google Scholar]
- 28.Chin W, Piuzzi F, Dimicoli I, Mons M. Probing the competition between secondary structures and local preferences in gas phase isolated peptide backbones. Physical Chemistry Chemical Physics. 2005;8:1033–1048. doi: 10.1039/b516245a. [DOI] [PubMed] [Google Scholar]
- 29.Gulyuz K, Wang D, Stedwell CN, Polfer NC. Gas-Phase Structure and Dissociation Chemistry of Protonated Tryptophan Elucidated by Infrared Multiple-Photon Dissociation Spectroscopy. Journal of Physical Chemistry Letters. 2011;2:299–304. Jr., W.K.M. [Google Scholar]
- 30.Sinhaa RK, Chiavarinoa B, Crestonia ME, Scuderib D, Fornarini S. Tyrosine nitration as evidenced by IRMPD spectroscopy. International Journal of Mass Spectrometry. 2011;308:209–216. [Google Scholar]
- 31.Thevis M, Schänzer W. Analytical approaches for the detection of emerging therapeutics and non-approved drugs in human doping controls. Journal of Pharmaceutical and Biomedical Analysis. 2014;101:66–83. doi: 10.1016/j.jpba.2014.05.020. [DOI] [PubMed] [Google Scholar]
- 32.Teale P, Scarth J, Hudson S. Impact of the emergence of designer drugs upon sports doping testing. Bioanalysis. 2012;4:71–88. doi: 10.4155/bio.11.291. [DOI] [PubMed] [Google Scholar]
- 33.Polfer NC, Oomens J, Suhai S, Paizs B. Spectroscopic and Theoretical Evidence for Oxazolone Ring Formation in Collision-Induced Dissociation of Peptides. Journal of the American Chemical Society. 2005;127:17154–17155. doi: 10.1021/ja056553x. [DOI] [PubMed] [Google Scholar]
- 34.Perkins BR, Chamot-Rooke J, Yoon SH, Gucinski AC, Somogyi A.r.d., Wysocki VH. Evidence of Diketopiperazine and Oxazolone Structures for HA b2+ Ion. Journal of the American Chemical Society. 2009;131:17528–17529. doi: 10.1021/ja9054542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stedwell CN, Patrick AL, Gulyuz K, Polfer NC. Screening for Phosphorylated and Nonphosphorylated Peptides by Infrared Photodissociation Spectroscopy. Analytical Chemistry. 2012;84:9907–9912. doi: 10.1021/ac3023058. [DOI] [PubMed] [Google Scholar]
- 36.Vernon MF, Krajnovich DJ, Kwok HS, Lisy JM, Shen YR, Lee YT. Infrared vibrational predissociation spectroscopy of water clusters by the crossed laser-molecular beam technique. Journal of Chemical Physics. 1982;77:47–57. [Google Scholar]
- 37.Stearns JA, Boyarkin OV, Rizzo TR. Spectroscopic signatures of gas-phase helices: Ac-Phe-(Ala)5-Lys-H+ and Ac-Phe-(Ala)10-Lys-H+ Journal of the American Chemical Society. 2007;129:13820–13821. doi: 10.1021/ja076507s. [DOI] [PubMed] [Google Scholar]
- 38.Stearns JA, Mercier S, Seaiby C, Guidi M, Boyarkin OV, Rizzo TR. Conformation-Specific Spectroscopy and Photodissociation of Cold, Protonated Tyrosine and Phenylalanine. Journal of the American Chemical Society. 2007;129:11814–11820. doi: 10.1021/ja0736010. [DOI] [PubMed] [Google Scholar]
- 39.Burke NL, Redwine JG, Dean JC, McLuckey SA, Zwier TS. UV and IR spectroscopy of cold protonated leucine enkephalin. International Journal of Mass Spectrometry. 2015;378:196–205. [Google Scholar]
- 40.Schinle F, Jacob Christoph R., Wolk Arron B., Greisch Jean-François, Vonderach Matthias, Weis Patrick, et al. Ion Mobility Spectrometry, Infrared Dissociation Spectroscopy, and ab Initio Computations toward Structural Characterization of the Deprotonated Leucine-Enkephalin Peptide Anion in the Gas Phase. Journal of Physical Chemistry A. 2014;118:8453–8463. doi: 10.1021/jp501772d. [DOI] [PubMed] [Google Scholar]
- 41.Boyarkin OV, Mercier SR, Kamariotis A, Rizzo TR. Electronic Spectroscopy of Cold, Protonated Tryptophan and Tyrosine. Journal of the American Chemical Society. 2006;128:2816–2817. doi: 10.1021/ja058383u. [DOI] [PubMed] [Google Scholar]
- 42.Wang X-B, Wang L-S. Development of a Low-Temperature Photoelectron Spectroscopy Instrument using an Electrospray Ion Source and a Cryogenically Controlled Ion Trap. Review of Scientific Instruments. 2008;79 doi: 10.1063/1.2957610. [DOI] [PubMed] [Google Scholar]
- 43.Wolk AB, Leavitt CM, Garand E, Johnson MA. Cryogenic Ion Chemistry and Spectroscopy. Accounts of Chemical Research. 2014;47 doi: 10.1021/ar400125a. [DOI] [PubMed] [Google Scholar]
- 44.Boyarkin OV, Kopysov V. Cryogenically cooled octupole ion trap for spectroscopy of biomolecular ions. Review of Scientific Instruments. 2014;85:033105. doi: 10.1063/1.4868178. [DOI] [PubMed] [Google Scholar]
- 45.Jašík J, Žabka J, Roithová J, Gerlich D. Infrared spectroscopy of trapped mo ecu ar dications below 4 K. International Journal of Mass Spectrometry. 2013;354–355:204–210. [Google Scholar]
- 46.Leavitt CM, Wolk AB, Fournier JA, Kamrath MZ, Garand E, Van Stipdonk MJ, et al. Isomer-Specific IR–IR Double Resonance Spectroscopy of D2-Tagged Protonated Dipeptides Prepared in a Cryogenic Ion Trap. Journal of Physical Chemistry Letters. 2012;3:1099–1105. doi: 10.1021/jz3003074. [DOI] [PubMed] [Google Scholar]
- 47.Hammer NI, Diken EG, Roscioli JR, Johnson MA, Myshakin EM, Jordan KD, et al. The vibrational predissociation spectra of the H5O2+·RGn (RG=Ar,Ne) clusters: Correlation of the solvent perturbations in the free OH and shared proton transitions of the Zundel ion. Journal of Chemical Physics. 2005;122:244301. doi: 10.1063/1.1927522. [DOI] [PubMed] [Google Scholar]
- 48.Jašík J, Roithová J. Infrared spectroscopy of CHCl2+ molecular dications. International Journal of Mass Spectrometry. 2015;377:109–115. [Google Scholar]
- 49.Roithová J, Gray A, Andris E, Jašík J, Gerlich D. Helium Tagging Infrared Photodissociation Spectroscopy of Reactive Ions. Accounts of Chemical Research. 2016 doi: 10.1021/acs.accounts.5b00489. [DOI] [PubMed] [Google Scholar]
- 50.Schwartz JC, Senko MW, Syka JEP. A two-dimensional quadrupole ion trap mass spectrometer. Journal of the American Society for Mass Spectrometry. 2002;13:659–669. doi: 10.1016/S1044-0305(02)00384-7. [DOI] [PubMed] [Google Scholar]
- 51.Douglas DJ, Frank AJ, Mao D. Linear ion traps in mass spectrometry. Mass Spectrometry Reviews. 2005;24:1–29. doi: 10.1002/mas.20004. [DOI] [PubMed] [Google Scholar]
- 52.Hager JW. A new linear ion trap mass spectrometer. Rapid Communication in Mass Spectrometry. 2002;16:512–526. doi: 10.1002/rcm.1020. [DOI] [PubMed] [Google Scholar]
- 53.Campbell JM, Collings BA, Douglas DJ. A new linear ion trap time-of-flight system with tandem mass spectrometry capabilities. Rapid Communication in Mass Spectrometry. 1998;12:1463–1474. doi: 10.1002/rcm.440. [DOI] [PubMed] [Google Scholar]
- 54.Cooper RJ, Heiles S, Williams ER. Effects of electronic structure on the hydration of PbNO3+ and SrNO3+ ion pairs. Physical Chemistry Chemical Physics. 2015;17:15963–15975. doi: 10.1039/c5cp01859e. [DOI] [PubMed] [Google Scholar]
- 55.DiTucci MJ, Heiles S, Williams ER. Role of Water in Stabilizing Ferricyanide Trianion and Ion-Induced Effects to the Hydrogen-Bonding Water Network at Long Distance. Journal of the American Chemical Society. 2015;137:1650–1657. doi: 10.1021/ja5119545. [DOI] [PubMed] [Google Scholar]
- 56.Marshall AG, Hendrickson CL. Fourier transform ion cyclotron resonance detection: principles and experimental configurations. International Journal of Mass Spectrometry. 2002;215:59–75. [Google Scholar]
- 57.Gerlich D. In: The study of cold collisions using ion guides and traps. Smith IWM, editor. Imperial College Press; London: 2008. [Google Scholar]
- 58.Olsen JV, Schwartz JC, Griep-Raming J, Nielsen ML, Damoc E, Denisov E, et al. A Dual Pressure Linear Ion Trap Orbitrap Instrument with Very High Sequencing Speed. Molecular & Cellular Proteomics. 2009;8:2759–2769. doi: 10.1074/mcp.M900375-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Redwine JG, Davis ZA, Burke NL, Oglesbee RA, McLuckey SA, Zwier TS. A novel ion trap based tandem mass spectrometer for the spectroscopic study of cold gas phase polyatomic ions. International Journal of Mass Spectrometry. 2013;348:9–14. [Google Scholar]