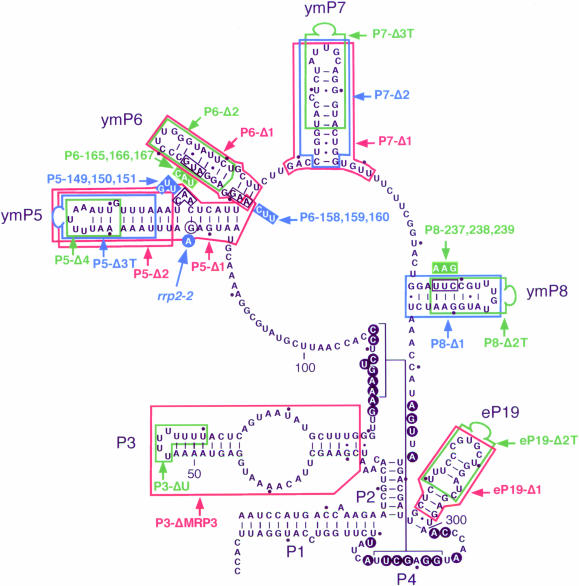
Figure 1.
Summary of mutant MRP RNA phenotypes. The secondary structure model for MRP RNA was determined as described previously (24). The P1, P2, P3 and P4 hairpins are found in all RNase P and RNase MRP RNA structures. Hairpin eP19 is found in eukaryotic RNase P and RNase MRP RNA, while hairpins ymP5, ymP6, ymP7 and ymP8 are found in RNase MRP RNA from the Saccharomycetaceae family [for details see (24)]. Deleted bases have been boxed; bases substituted are also boxed with the replacement sequence indicated next to the box. Deletion mutants with an inserted tetra-loop (CGAAAG) are indicated by the suffix T and a loop at the end of the boxed bases. Mutations indicated in green have no significant effect on growth. Mutations in blue have a conditional phenotype, not growing at high temperature and, with the exception of P6-158,159,160, growing very slowly at lower temperatures. Mutations in red are lethal. Bases conserved in all currently known MRP sequences are indicated with black circles. The deletions in hairpin P3 have been described previously (26). The single base change in hairpin ymP5 (rrp2-2) has also been described before (4).