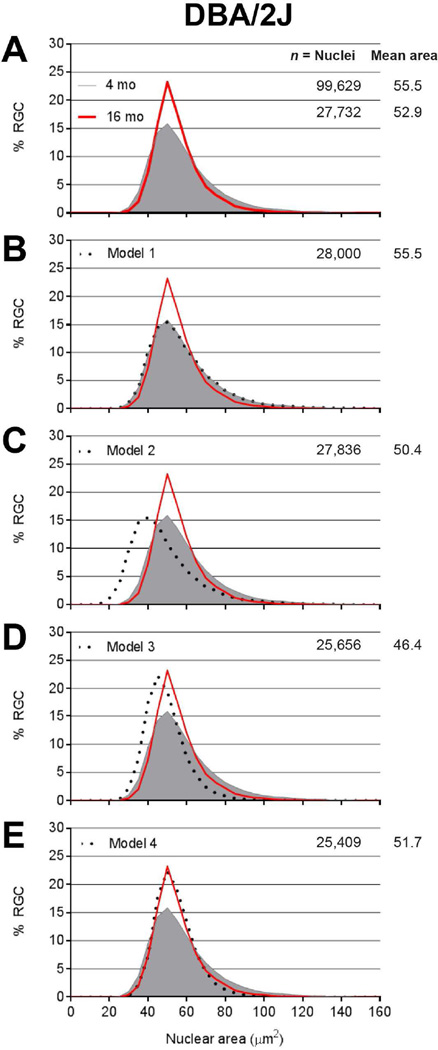
Figure 6. Subtle enlargement of all retinal ganglion cell nuclei and preferential death of cells with the largest nuclei in DBA/2J mice with glaucoma.
Frequency distribution plots of nuclear size for pre-disease 4-month-old DBA/2J (solid gray), glaucomatous 16-month old DBA/2J (red), and various models (dotted black) tested for whether a manipulation of the 4-month-old dataset could cause it to resemble the 16-month-old dataset. (A) Observed data for DBA/2J glaucoma showing reductions in cell number and nuclear area, with contractions of the distribution indicating a preferential loss of cells with largest and smallest nuclei. Asterisk, p=1.2E−24, Mann-Whitney Test. (B) Model 1 showing application of a random 72% loss of all nuclei. Note that the frequency distribution plot of the model completely overlaps that of pre-disease mice, indicating poor performance of the model. (C) Model 2 showing application of a uniform 5 µm2 atrophy to all nuclei followed by a random 72% loss of all nuclei. Note that that the frequency distribution plot of the model does not resemble that of the glaucomatous mice and that the mean nucleus size is too small, indicating poor performance of the model. (D) Model 3 showing a size-dependent loss of nuclei, with half of the risk fixed at 50% and the other half continuously increasing according to size rank amongst all retinal ganglion cell nuclei in the population. Note that the frequency distribution plot of the model does not resemble that of the glaucomatous mice and that the mean nucleus size is too small, indicating poor performance of the model. (E) Model 4 showing the same size-dependent loss of nuclei as Model 3, but with an accompanying and uniform 5 µm2 enlargement of all remaining nuclei. Note that the frequency distribution plot of the model closely resembles that of the glaucomatous mice, indicating good performance of the model.