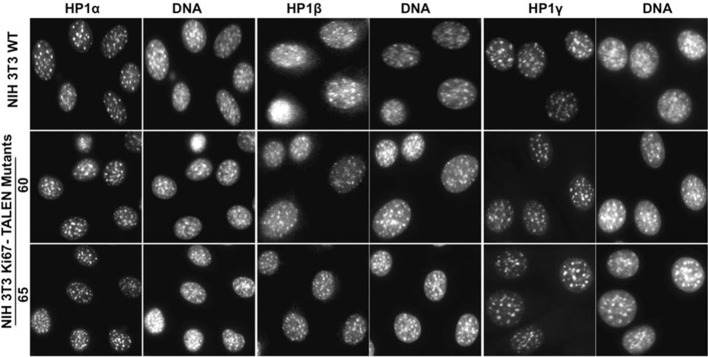
Figure 10. Ki-67 controls heterochromatin organisation.
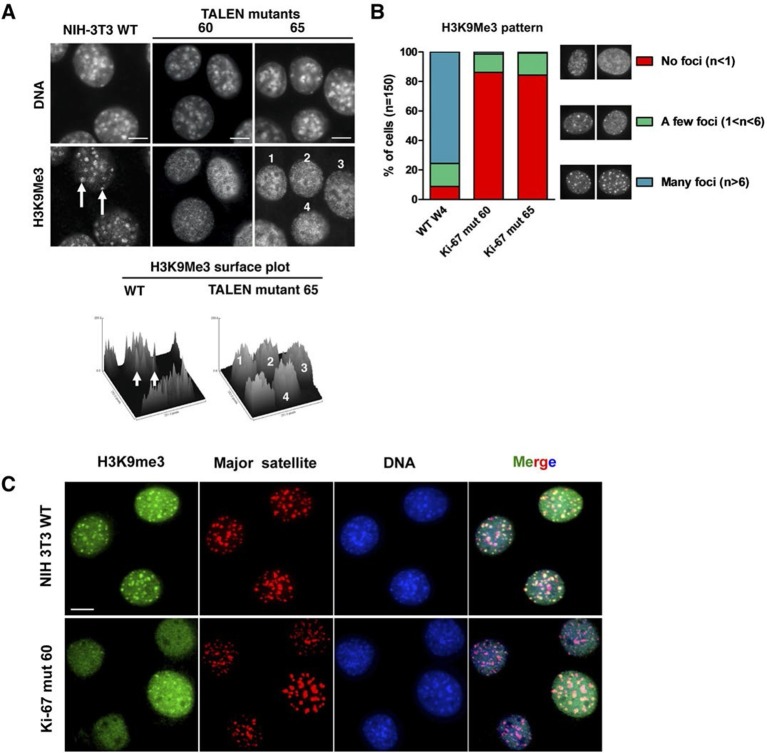
(A) Top, immunofluorescence analysis of H3K9me3 in mouse NIH-3T3 WT and Mki67 TALEN mutant clones 60 and 65. Bars, 5 µm. Below: graphs showing quantification of pixel intensity scans for H3K9me3. (B) Quantification of H3K9Me3 patterns in WT and Ki-67 mutant clones. (C) Immunofluorescence of H3K9Me3, FISH of major satellite DNA and DAPI staining in WT W4 and Ki-67 mutant clone 60. Bar, 10 µm.
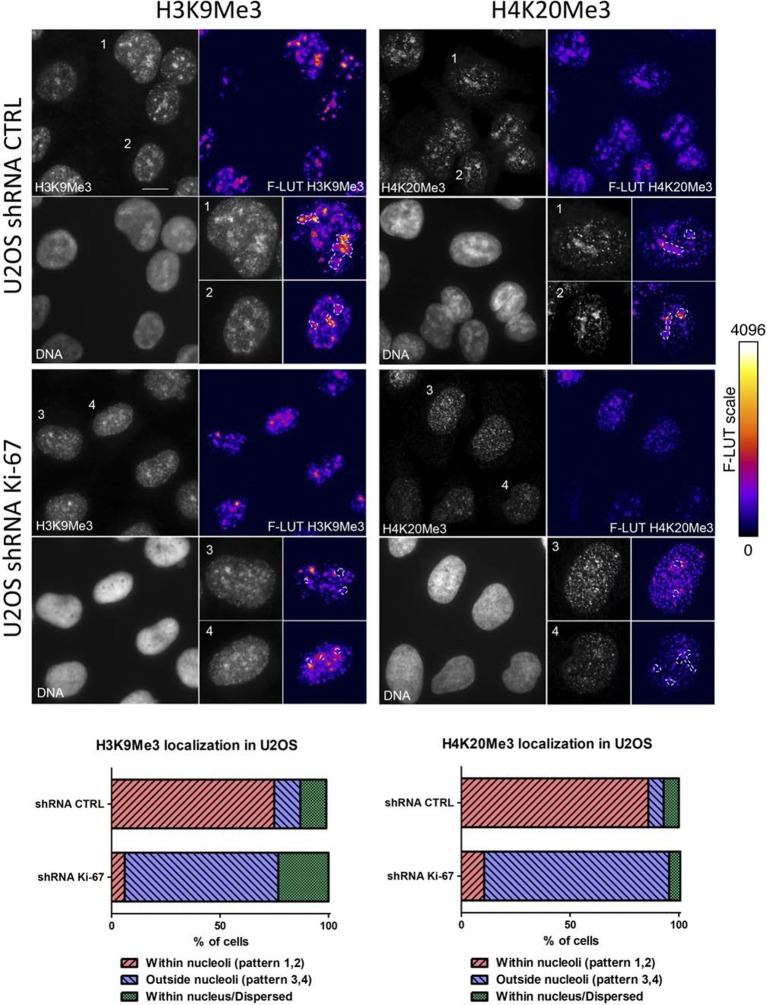
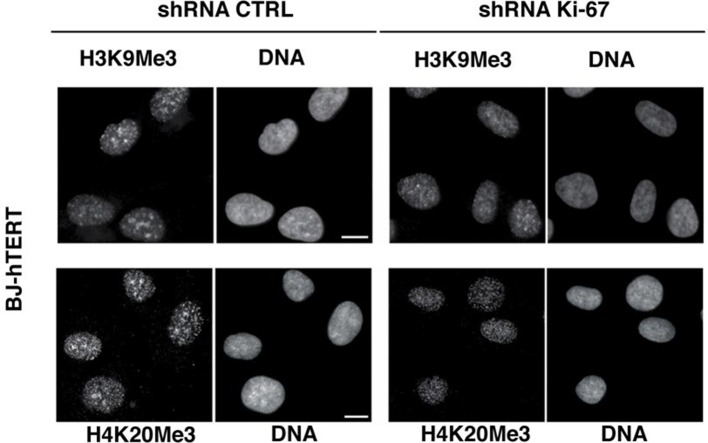
Figure 10—figure supplement 1. Heterochromatic histone mark localisation requires Ki-67.
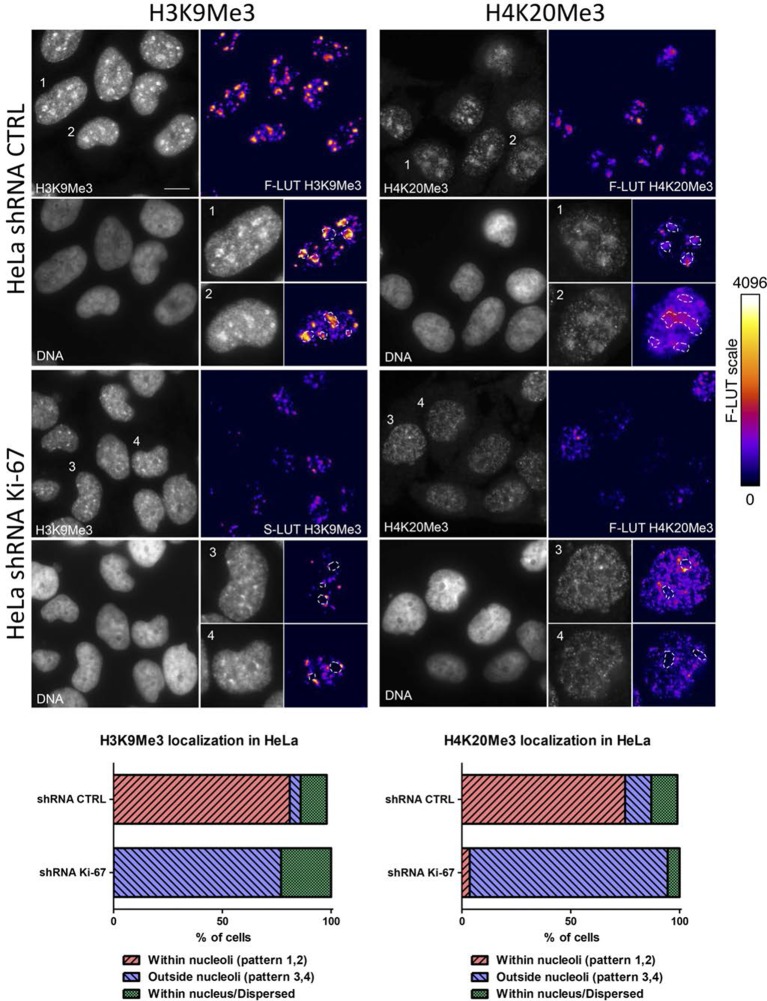
Figure 10—figure supplement 2. Heterochromatic histone mark localisation requires Ki-67.
Figure 10—figure supplement 3. Heterochromatic histone mark localisation requires Ki-67.
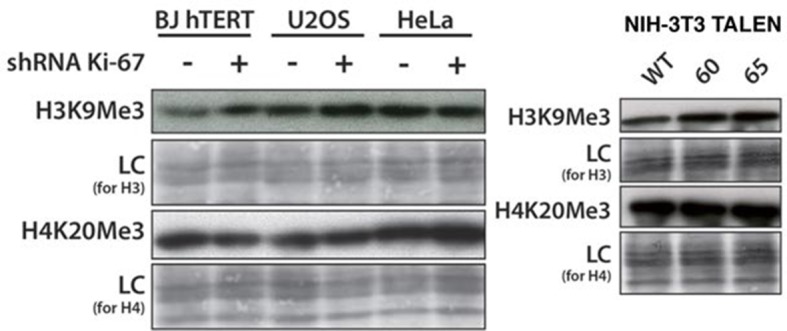
Figure 10—figure supplement 4. Overall heterochromatic histone mark levels do not change upon Ki-67 knockdown.