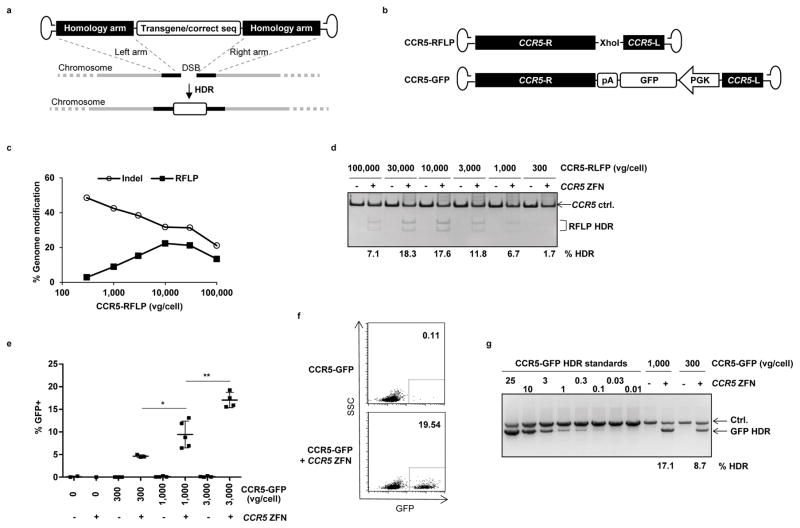
Figure 2. Combination of ZFN mRNA and AAV6 vectors promotes high levels of site-specific genome editing at the CCR5 locus in HSPCs.
(a) Schematic showing use of AAV vector as a template for homology directed repair (HDR) of a double-strand break (DSB), as induced by target-specific nucleases. (b) Schematic of AAV vector genomes containing CCR5 homology donors. R and L refer to CCR5 genomic sequences, comprising 1431 and 473 bp respectively, and inserted in antisense orientation when compared to the AAV genome20. Vector CCR5-RFLP contains an additional Xho1 restriction site and vector CCR5-GFP contains a promoter GFP cassette with a polyadenylation (pA) sequence. (c) Mobilized blood CD34+ HSPCs were transduced with AAV6 vectors carrying the CCR5-RFLP donor at indicated doses for 16 hours, then electroporated with CCR5 ZFN mRNA (120μg/ml). Cells were analyzed 3–5 days post-electroporation by deep sequencing to measure the efficiency of genome modification (% indels and site-specific RFLP insertions). Results from one representative of 3 experiments using 3 different HSPC donors are shown. (d) Dose-dependent insertion of XhoI site at CCR5, confirmed by RFLP analysis. One representative experiment is shown, with % HDR quantitation for any sample greater than background. (e) Mobilized blood HSPCs were treated as described, but using CCR5-GFP donor vectors, with and without CCR5 ZFN mRNA electroporation. Cells were collected 3–6 days post-transduction and analyzed by flow cytometry for % GFP+. Results were combined from 5 experiments using 4 different donors and show mean +/− SD. * p<0.05, ** p<0.01, unpaired t-test. (f) Flow cytometry plots from one representative experiment using 3,000 vg/cell CCR5-GFP donor, at 6 days post-electroporation. (g) Confirmation of targeted integration of GFP expression cassette at the CCR5 locus by semi-quantitative In-Out PCR, for one representative experiment. Ctrl. is a PCR that serves as a genomic DNA loading control. The % HDR-mediated insertion of GFP was estimated following normalization and by comparison to standards, and numbers are shown for any samples greater than background. Uncropped images of all gels in this figure are available in Supplemental Figure 10.