Fig. 1.
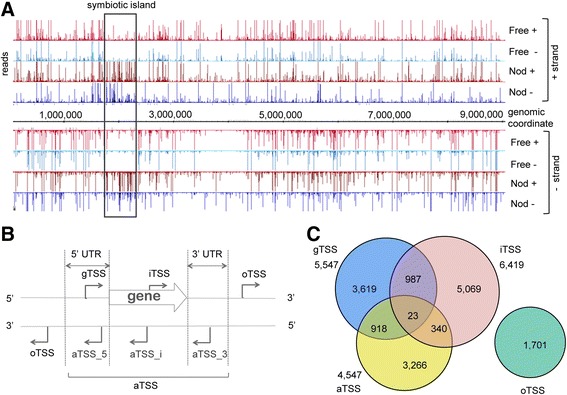
Differential RNA-seq of B. japonicum USDA 110 and TSSs mapping. a cDNA reads mapped to the 9.1 Mb genome. RNA was isolated from exponentially growing, free-living cells (Free) in liquid cultures and from nodules (Nod). RNA samples were treated (+) or not treated (–) with TEX. As expected, there are more reads in the Nod libraries mapped to the symbiotic island compared to the Free libraries (framed). All libraries were adjusted to the same scale (0 to 300 reads). b Categories for TSS annotation based on the genomic context: gTSS, TSS of an annotated gene; iTSS, internal TSS (located in an annotated gene) in sense orientation; aTSS, antisense TSS (aTSSs are divided in the following sub-categories: aTSS_5, aTSS located in a region defined as 5′ UTR; aTSS_i, internal aTSS located inside an annotated gene; aTSS_3, aTSS located in a region defined as 3′ UTR); oTSS, orphan TSS, all TSSs which do not fall in the categories mentioned above. c Venn diagram, showing distribution of the TSSs among the categories gTSS, iTSS, aTSS and oTSS
