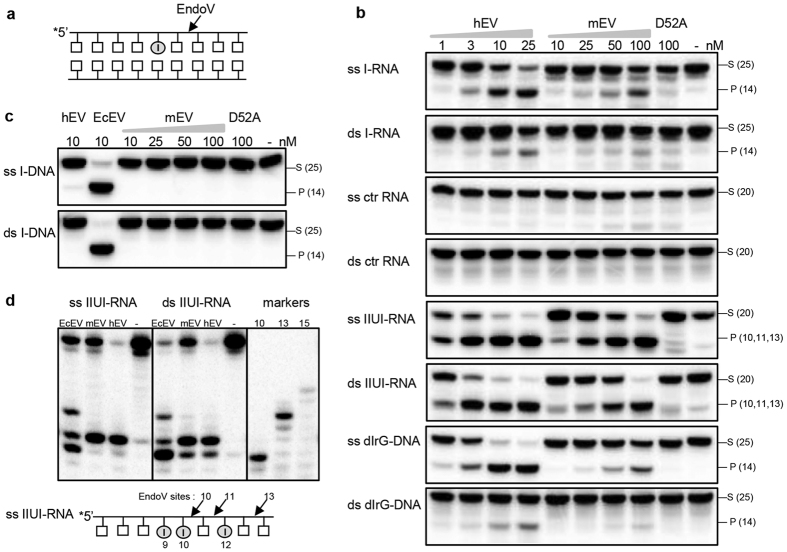
Figure 1. Processing of inosine by EndoV enzymes.
(a) Schematic illustration of EndoV cleavage of the second phosphodiester bond 3′ to an inosine residue. (b) Increasing amounts of human EndoV (hEV;1–25 nM) and mouse EndoV (mEV; 10–100 nM) were incubated with 1 nM RNA or DNA substrates: ss/ds I-RNA, ss/ds ctr-RNA, ss/ds IIUI-RNA, ss/ds dIrG-DNA and (c) ss/ds I-DNA at 37 °C for 30 minutes using standard reaction buffer. 100 nM of the site specific mEndoV mutant D52A was included in all assays. Cleavage products were analyzed by 20% denaturing PAGE and visualized by phosphorimaging. (d) EndoV cleavage products for the IIUI-RNA substrates were run on 20% sequencing gels alongside with 10, 13 and 15 mer RNA markers. The illustration shows potential EndoV cleavage sites on the IIUI-RNA substrate. Abbreviations: - = no enzyme added, S = substrate, P = cleaved product, ss = single-stranded, ds = double-stranded, EcEV = E. coli EndoV. Enzyme amounts are shown in nM and sizes of RNA/DNA substrates and products are indicated in parentheses.