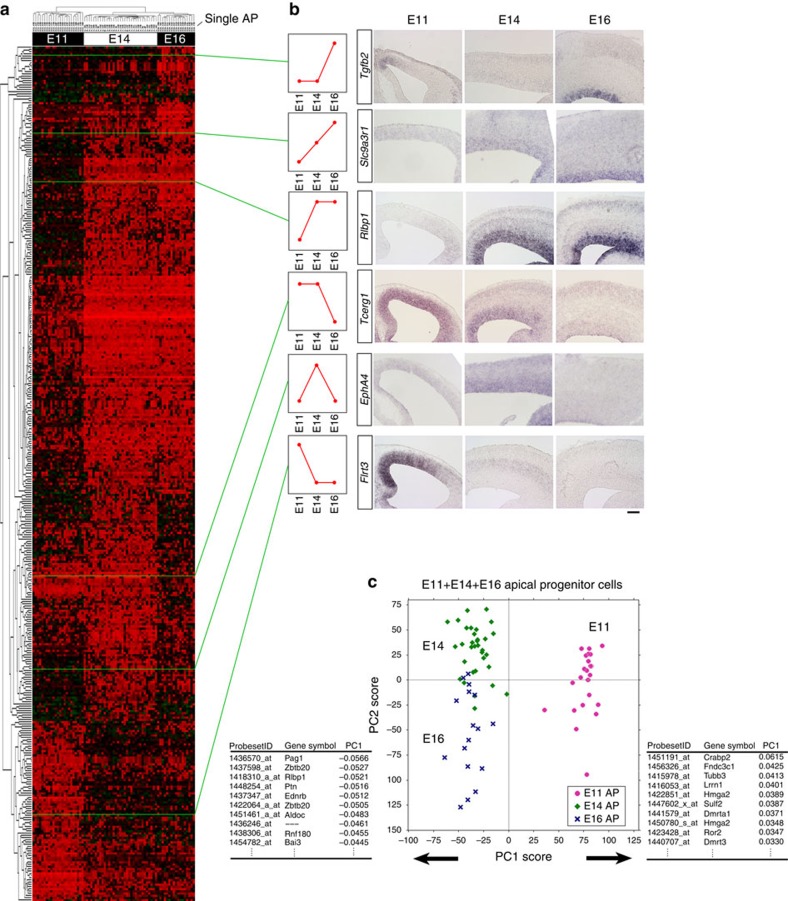
Figure 2. Temporal change in gene expression in APs.
(a) Two-way cluster analysis of genes differentially expressed among E11, E14 and E16 single-cell cDNAs of the APs (384 probe sets, Supplementary Data 1, selected by one-way ANOVA). Examples of genes that exhibited typical expression patterns in E11, E14 and E16 cerebrum are shown in b or Supplementary Figs 5 and 6. The ‘medial>lateral' genes exhibited an ‘E11>E14' trend, and the ‘medial<lateral' genes exhibited an ‘E11<E14' trend (Supplementary Fig. 7). Scale Bar, 100 μm (c) Global gene-expression patterns of E11 APs are very different from those of E14/E16 APs. PCA was performed on microarray data from single-cell cDNAs of all APs (total N=73: mixture of E11, N=23; E14, N=33; E16, N=17 single cells; 17192 probe sets). Each symbol indicates one cell. PC1, the most representative axis for the gene-expression variation among the AP population, is determined by the difference between E11 and E14/E16 cells. The lists below indicate the top 10 genes that positively or negatively influence PC1. Proportion of variance: 0.0436 (PC1) and 0.0319 (PC2). See also Supplementary Figs 5–9 and Supplementary Data 1.