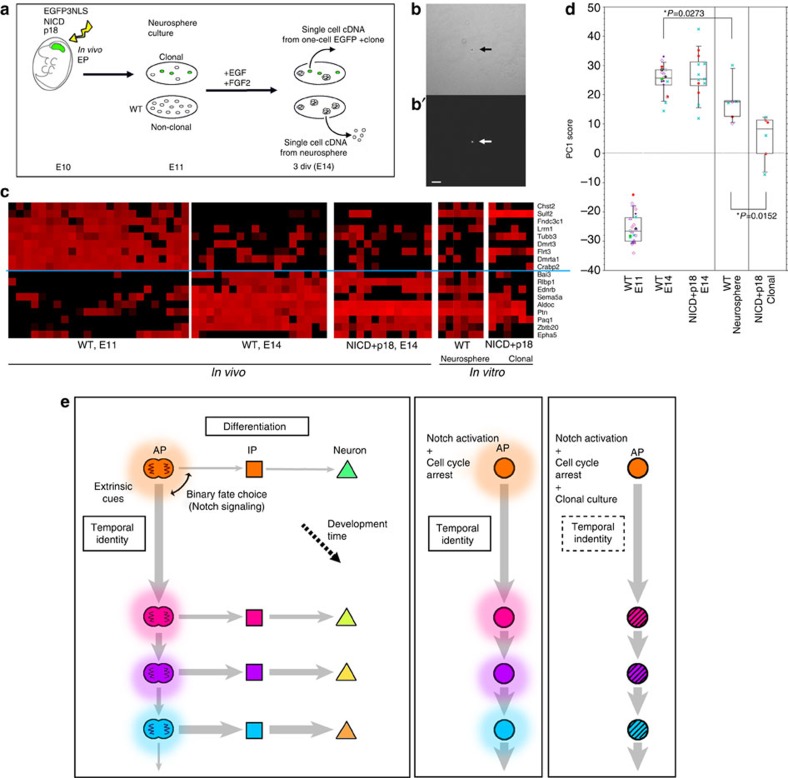
Figure 7. Temporal change of gene expression in APs is partly cell autonomous.
(a) Experimental design. (b) Single EGFP+ cell (arrow) formed one-cell clone at 3 div in clonal culture. b, Bright field; b′, EGFP fluorescence. Scale bar, 50 μm. (c) Expression levels of 18 temporal-axis genes in single APs examined by qPCR. Levels range from high (red) to low/undetectable (black) in these heat maps. Each column indicates a single AP (N=24, wild-type E11 APs; N=18, wild-type E14 APs; N=13, E14 APs electroporated with NICD/p18 at E11 (identical to the data shown in Fig. 4); N=6, APs from neurospheres; N=6, NICD+p18 co-expressing APs from one-cell clones. NICD+p18 co-expressing APs from one-cell clones were Egfp+/Ttyh1+/Sox2+/Hes5+/Tbr2−/Ki67−, as determined by qPCR. (d) PC1 scores of wild-type APs at E11 and E14, NICD/p18 co-expressing APs at E14 (identical to the data shown in Fig. 4), APs from neurospheres, and NICD+p18 co-expressing APs from one-cell clones, which were calculated using Component 1 obtained from PCA of wild-type APs (Fig. 4e). Note that the PC1 scores for the neurosphere-derived APs differ significantly from those for the wild-type E14 APs (P=0.0273, Mann–Whitney U test). See also Supplementary Fig. 16, which shows the characterization of the microarray data from the single neurosphere-derived APs. (e) Temporal patterns of cortical progenitor cells (model). Transition of temporal identity of APs, which occurs gradually over the course of development, cannot be stopped by constitutive Notch activation or cell-cycle arrest in vivo. This transition of temporal identity includes both the transition in division patterns and transition in laminar fate potential of APs. Clonal culture of cell-cycle-arrested APs partly impairs transition of temporal gene expression, suggesting that the transition in temporal identity is regulated by both cell-autonomous and non-cell-autonomous mechanisms.