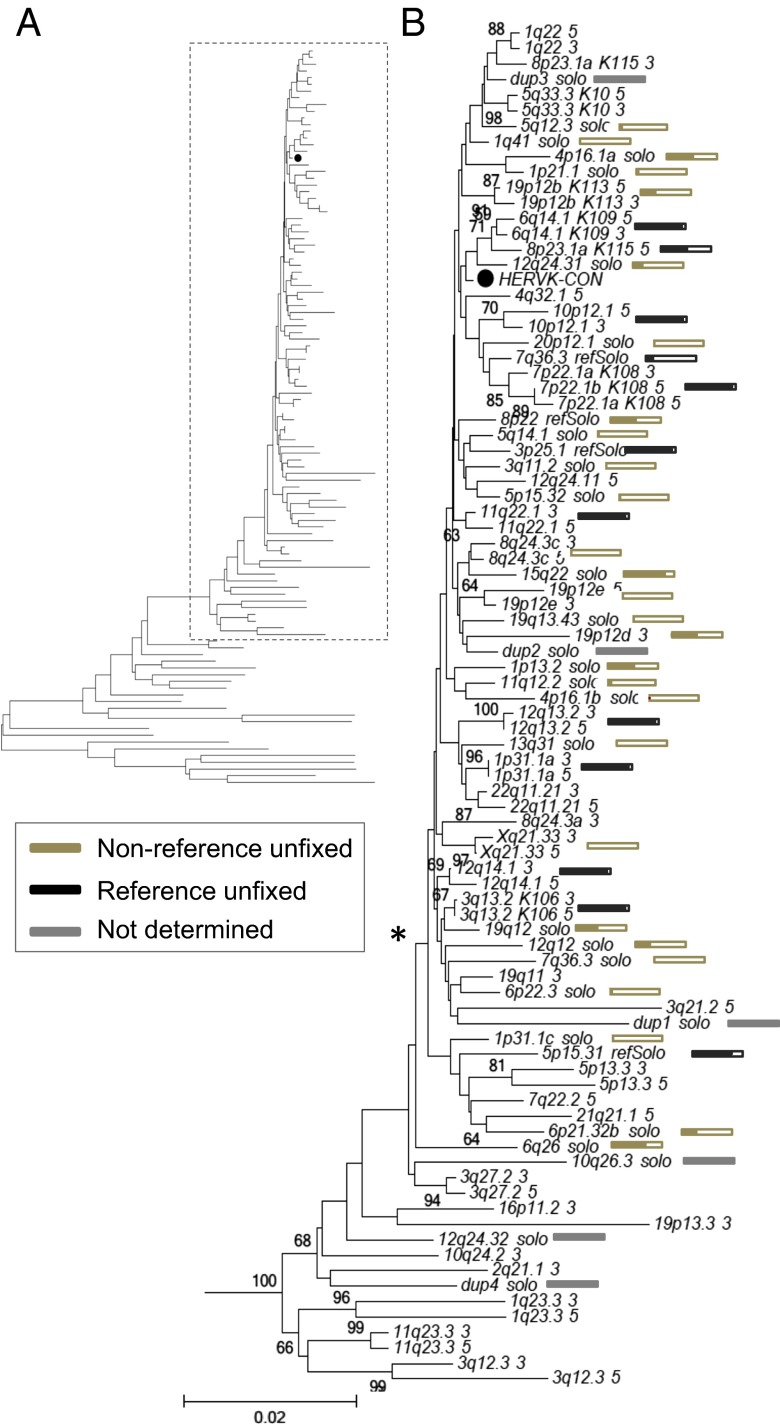
Fig. 3.
Phylogenetic construction of HML-2 LTRs within humans. (A) Neighbor-joining tree was constructed based on the aligned nucleotide sequences corresponding to HML-2 LTRs from the LTR5Hs group, specifically including those nucleotide sequences considered to be human-specific and/or polymorphic. The LTRs were extracted from (i) all reference HML-2 proviruses previously inferred as belonging to the LTR5Hs HML-2 subgroup [as reported by Subramanian et al. (11)], (ii) unfixed reference solo-LTRs [as reported by Belshaw et al. (5), and (iii) unfixed nonreference insertions as reported here. Both 5′ and 3′ LTRs were used for full-length insertions, when present. The closed circle (●) indicates the taxon corresponding to the HERV-KCON LTR within the tree. Classic nomenclature has been included in taxon names for the better studied insertions: K113 (19p12b), K108 (7p22.1), K115 (8p23.1a), K106 (3q13.2), and K109 (6q14.1). (B) Detailed view of branches representing unfixed HML-2 insertions. Individual HML-2 loci are indicated for each branch as follows: the cytoband followed by a 5′ or 3′ for the 5′ or 3′ LTRs from full-length insertions, solo for nonreference unfixed solo-LTR insertions, or refSolo for reference unfixed loci. An asterisk is used to indicate the position of the clade containing the majority of unfixed insertions. Boxes are used to indicate estimated allele frequencies for each unfixed insertion at the end of each respective branch. The filled area within each box is shown as proportional to the estimated frequency of the insertion in all samples; the derived values are provided in Dataset S3. Gold and black boxes are used to represent nonreference and reference unfixed insertions, and gray bars indicate the elements for which the frequency could not be determined.