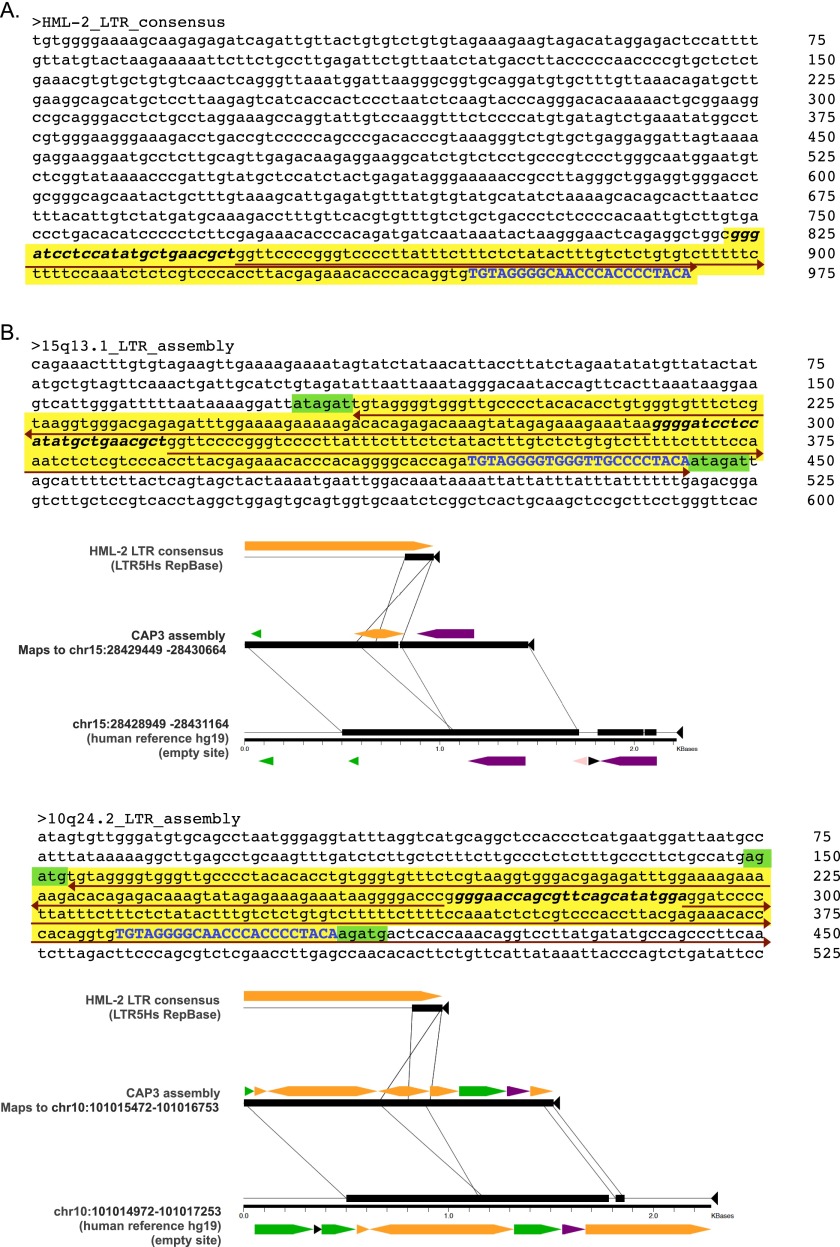
Fig. S2.
Assembled loci with unusual structure. Detailed nucleotide structure of the assembled contigs at 15q13.1 and 10q24.2. (A) Consensus LTR from the most recent HERV-K HML-2 insertions corresponds to the 968-bp LTR5_Hs consensus (RepBase release 11.03). The last 146 bp (yellow shading) are detected in two of our assembled structures; specific regions are labeled to indicate structural similarities with the two assembled elements as follows. (B) Nucleotide sequences and Miropeats alignments derived in local read assemblies at the 15q13.1 (Upper) and 10q24.2 (Lower) loci. Shading is used to discriminate LTR-derived portions from flanking genomic sequence. The yellow shading indicates an LTR5_Hs-matching sequence that assembles as an inverted repeat (red arrows) with a central unique portion (bolded and italicized in all three sequences) at those sites. The presence of a short stretch of the LTR-derived portion that is present in the hg19 reference is shown in blue, and the putative TSDs of 6 bp and 5 bp, respectively, are shaded in green. Block arrows indicate RepeatMasker annotated repeats within each aligned segment. DNA repeats are shown in pink, LTRs are shown in orange, long interspersed elements (LINEs) are shown in green, short interspersed elements (SINEs) are shown in purple.