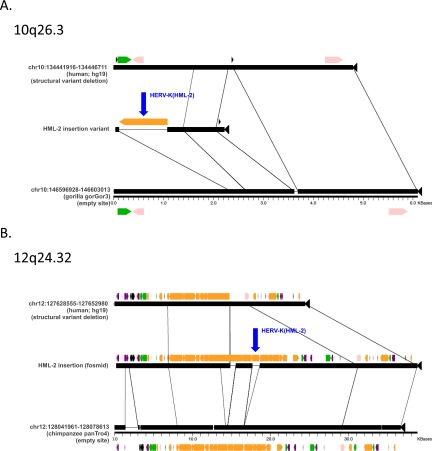
Fig. S3.
Insertions located within genomic structural variants. Aligned sequence segments were generated using the program Miropeats (58) for HML-2 insertions in encompassing deletions that exist as structural variants in humans, located within 10q26.3 (A) and 12q24.32 (B) relative to the hg19 reference. Junction sequences corresponding to these insertions were recovered in BLAST searches of the NCBI Trace Archive; the adjacent flanking sequence maps to putative preinsertion sites within nonhuman primate reference genomes (gorilla or chimpanzee indicated as appropriate, including positions mapping to the preinsertion alleles in those references). The alignments corresponding to either insertion are arranged to indicate the absence of the insertion in the hg19 segment (Top), the presence of the insertion within the validated variant region (Middle), and the homologous sequence from the reference to which the preinsertion site could be mapped (Bottom). Lines are used to indicate segments of homology between the aligned segments. The validated LTR at 10q26.3 was sequenced in this study (HG00449), and the LTR at 12q24.32 was taken from a finished fosmid clone from Kidd et al. (43) (NIH CloneDB: AC1945745.1). Repeats are colored as in Fig. S2. The LTRs corresponding to the identified HML-2 insertions at 10q26.3 and 12q23.32 are labeled in blue with arrows.