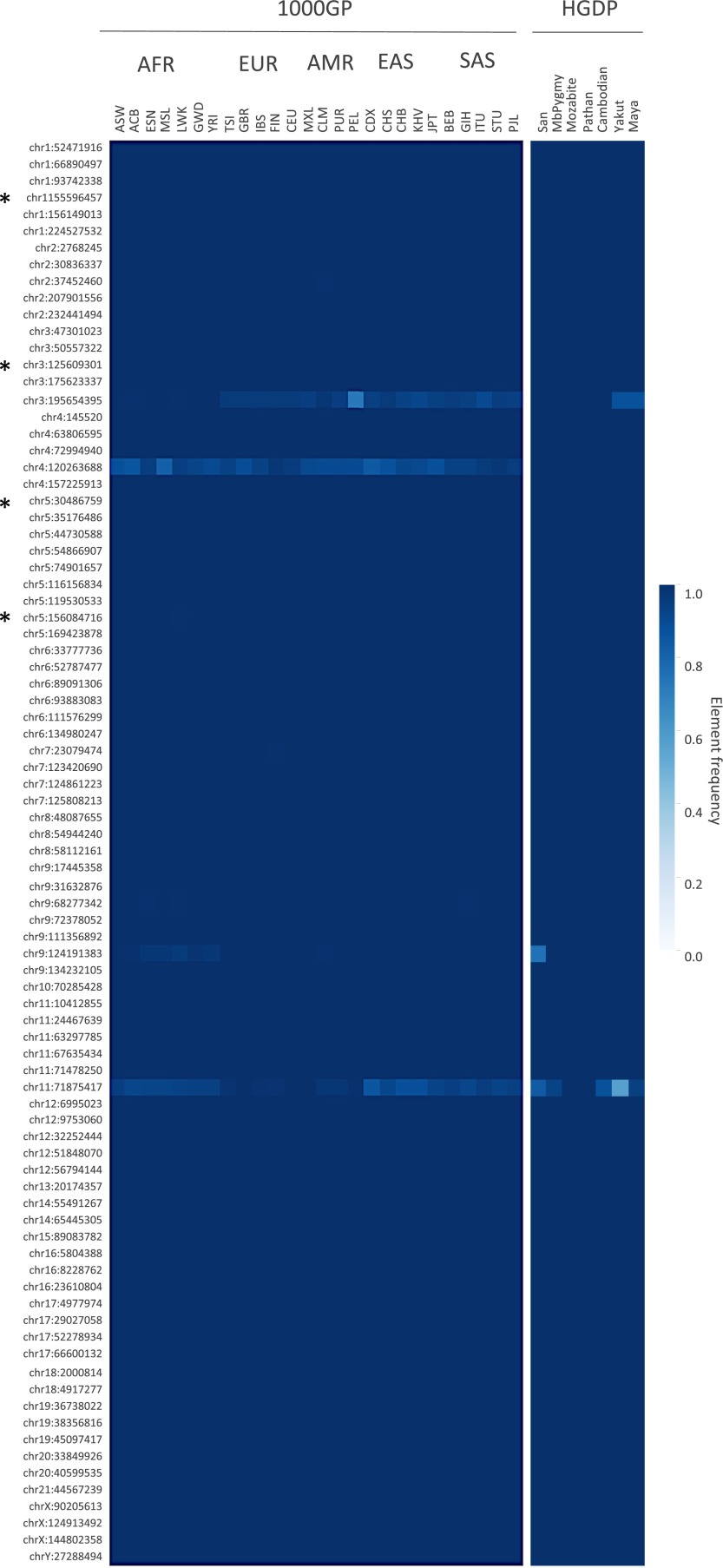
Fig. S4.
Inferred frequencies of additional human-specific HML-2 insertions in humans. A total of 85 additional HML-2 loci were subjected to in silico genotyping based on previous reports of human-specific HML-2 elements from Belshaw et al. (5) and Subramanian et al. (11). Of these loci, 81 insertions are present as solo-LTRs and four (indicated to the left by an asterisk) are present as proviruses in the hg19 reference genome, excluding insertions with unmatched TSDs and requiring the presence of cognate LTRs for full-length proviruses. Three solo-LTRs corresponded to duplicated loci and were excluded (chr8_91696140, chrY_25039185, and chrY_26672934). Genotypes were inferred by remapping Illumina reads to reconstructed reference (i.e., insertion) or alternate (i.e., empty) alleles for each site. Population allele frequencies were calculated as the total number of insertion alleles divided by total alleles and shown by heat map. The 1KGP and HGDP populations are labeled above (also refer to Dataset S1). The locus of each of the unfixed HML-2 loci is labeled to the left according to its chromosomal position.