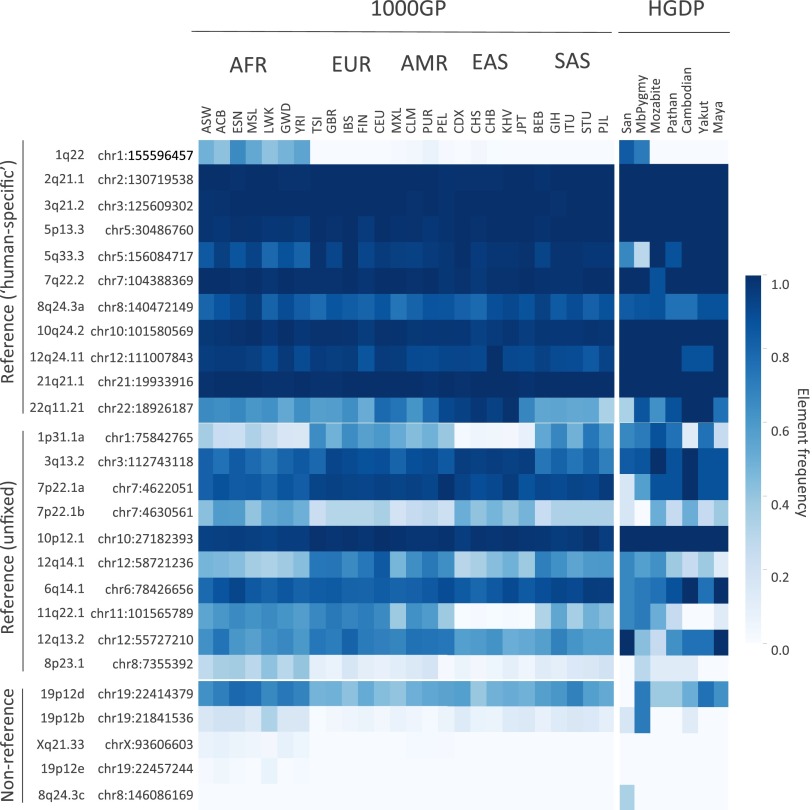
Fig. S6.
Presence of full-length unfixed HML-2 proviruses in the human genome. The presence of provirus (2-LTR) insertions was inferred based on counting of unique k-mers identified from the full nucleotide sequences corresponding to unfixed full-length insertions. Cytoband information and chromosomal positions corresponding to each locus are shown to the left. Sample carrier frequencies are represented as a heat map as shown by the color legend to the right. Samples are labeled by population according to the 1KGP (1000GP) or HGDP dataset (also refer to Dataset S1). The estimated distribution of reference unfixed HML-2 with confirmed 2-LTR proviruses [from loci reported by Subramanian et al. (11)] and the estimated distribution of nonreference HML-2 2-LTR insertions reported here are shown. AFR, African; AMR, Admixed American; EAS, East Asian; EUR, European; SAS, South Asian.