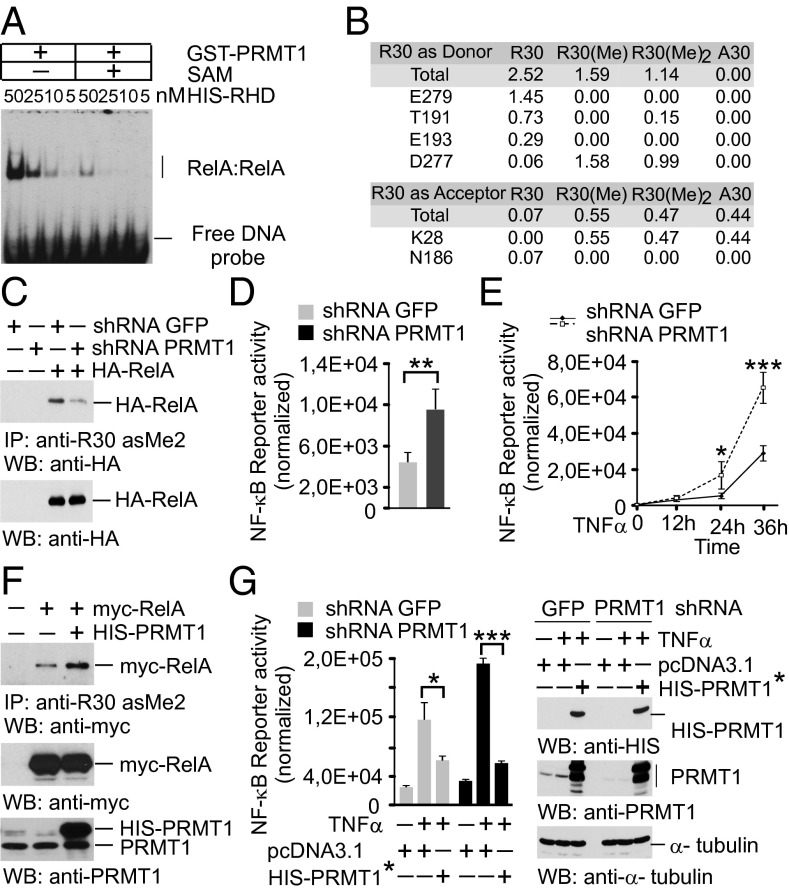
Fig. 4.
RelA methylation by PRMT1 inhibits the DNA-binding and transcriptional activity of NF-κB. (A) EMSA with nonmethylated and in vitro methylated RelA was performed as shown in Fig. 3A. (B) Hydrogen-bonding occupancy of nonmethylated, monomethylated, and asymmetrically dimethylated R30 and A30 in molecular dynamics simulations. (C) HEK 293T cells expressing shRNAs were transfected with control vector or an HA-RelA construct. RelA methylation was analyzed as in Fig. 2C, followed by immunoblotting using anti-HA antibody. (D) Control and PRMT1 knockdown HEK 293T cells were transfected with 2×(κB)-luciferase reporter. Luciferase activity was measured in cell lysates and normalized to protein content. (E) Cells were transfected with the NF-κB reporter and stimulated with 20 ng/mL TNFα for 40 min. Luciferase activity was measured at the indicated time points. (F) HEK 293T cells were cotransfected with myc-RelA and His-PRMT1–expressing plasmids. R30 methylation was analyzed as above. (G) Cells were transfected with control or shRNA-resistant His-PRMT1* constructs. (Left) Luciferase assays were performed as described in E. (Right) The expression of His-PRMT1 was controlled by immunoblotting. Values are means ± SD of triplicates. *P < 0.05; **P < 0.01; ***P < 0.001.