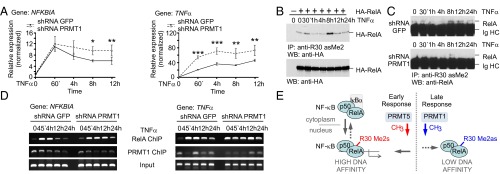
Fig. 5.
PRMT1 regulates NF-κB target genes in response to TNFα. (A) NFKBIA and TNFα gene-specific mRNAs were quantified at the indicated time points after TNFα stimulation using qRT-PCR and the comparative ddCt method. Values are means ± SD of at least three experiments. *P < 0.05; **P < 0.01; ***P < 0.001. (B and C) The time course of methylation of ectopic (B) or endogenous RelA (C) in response to TNFα was analyzed as in Fig. 2C, followed by immunoblotting using anti-HA or anti-RelA (C20) antibodies, respectively. (D) ChIP analysis of promoter occupancy by RelA and PRMT1 in TNFα-treated control and PRMT1 knockdown cells. (E) Model for PRMT1-mediated regulation of NF-κB. In addition to IκB-mediated repression, NF-κB is negatively regulated by methylation of the RelA subunit that is catalyzed by PRMT1. Asymmetric R30 dimethylation in RelA inhibits the transcription of TNFα-induced genes by reducing the DNA binding of NF-κB. Symmetric R30 dimethylation, reported previously by Wei et al. (16), is seen as an NF-κB–activating mark. It is postulated that symmetric and asymmetric RelA dimethylation may represent a specific on/off switch mechanism modulating cytokine-induced NF-κB responses.