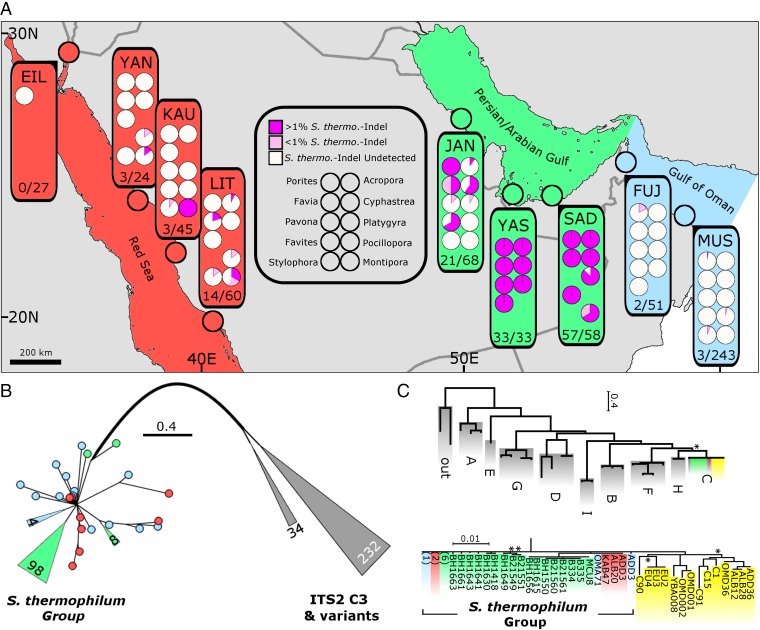
Fig. 1.
Identification and phylogenetic resolution of the S. thermophilum group. Colors denote sampling locations according to map coloration with yellow denoting non-S. thermophilum group clade C symbionts. (A) Sampling sites (three letter abbreviations; e.g., JAN) are detailed in Table S1. The presence of S. thermophilum group symbionts within the 10 most sampled genera are presented categorically (undetected, <1% and >1% S. thermo.-indel). Regional summaries (*)/(*) represent the proportion of samples in which S. thermophilum group symbionts were detected (Tables S1 and S2). The map was created as detailed in Hume et al., 2015 (13). (B) PsbAncr estimated phylogeny of S. thermophilum group samples created through the addition of samples collected in the Gulf of Oman and the Red Sea (sequenced as part of this study) to psbAncr alignments previously constructed (15). Symbionts within the gray branches are ITS2 type C3 and closely related ITS2 types from the Atlantic and Indo-Pacific (17). Wedges and circles represent collapsed groups and individual samples, respectively. Lengths of collapsed branches are drawn to scale with number of contained sequences indicated. (C) Supermatrix Bayesian estimation of phylogeny using six additional genetic markers (Fig. S3 and Table S5) on a selection of cnidarian-harbored Symbiodinium samples collected in the PAG, the Gulf of Oman, the Red Sea, as well as several reference locations (see Methods for further details). The full collapsed tree with clade C subtree is expanded below. Numbers in parentheses represent multiple sequences resolving at the same position and are colored according to collection site and sequence type (Gulf of Oman, blue; PAG, green; Red Sea, red; reference sequences, yellow). Gray groupings contain reference Symbiodinium from clades A–I as well as two outgroups (G. simplex and P. glacialis) (Table S5). Nodes supported with posterior probability (PP) above 0.8 are not displayed. Nodes with supports below 0.8 are marked with an asterisk (*).