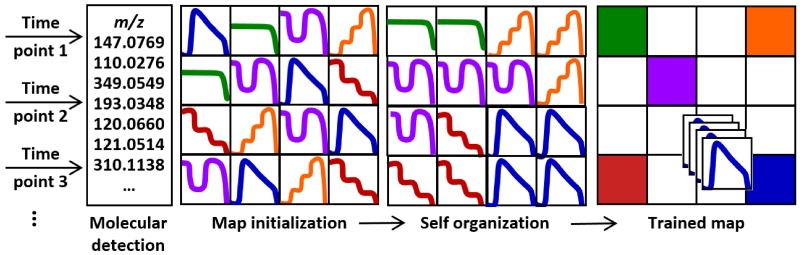
Figure 2.
A conceptual workflow for the self-organization of high dimensional data using MEDI. A series of untargeted experiments are performed for which a separate tile is constructed for each molecular feature as a function of abundance extracted for each experimental condition, such as different time points in longitudinal exposure. After molecular detection, the map initialization step randomly generates a map with intensity profiles indicative of the data. During the self organization training process, intensity profiles (or tiles) are grouped based on similarities in an iterative process until they are matched to their closest matching profile. Once the training phase is complete and a grid location determined, heat maps are generated for each sample based upon the intensity of the seeded features within that sample. These self-organized heat maps can then be averaged and/or differentially analyzed to distinguish regions of interest that differ among samples.