Figure 6. Mechanisms by which DYRK2 regulates the proteasome.
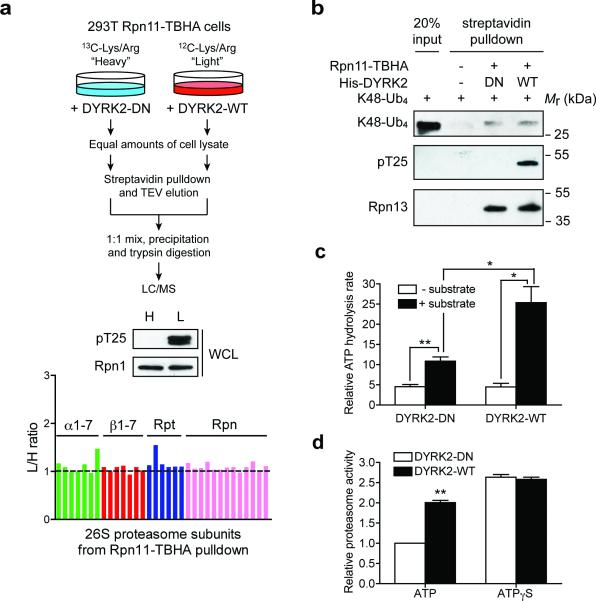
(a) DYRK2 does not affect 26S proteasome assembly. 293T Rpn11-TBHA cells labeled with “heavy (H)” or “light (L)” isotopes were transfected with inactive or WT DYRK2, respectively. Proteasomes were isolated from these cells and analyzed by quantitative mass spectrometry (top). DYRK2-WT strongly phosphorylated Rpt3-T25 (middle), without affecting the relative abundance of each subunit in the purified 26S proteasomes (bottom).
(b) In vitro binding of K48-linked tetraubiquitin (K48-Ub4) to the proteasome. Control 293T cells and 293T Rpn11-TBHA cells were subjected to streptavidin pulldown. The purified proteasome was treated with DYRK2-WT or DN in vitro and then incubated with K48-Ub4. Proteasome-bound K48-Ub4 was probed with anti-ubiquitin antibody. Western blot of the ubiquitin receptor Rpn13/ADRM1 shows equal amounts of proteasome in the DYRK2-treated samples.
(c) Affinity-purified 26S proteasomes (~ 1 μg) was treated with DYRK2-WT or D275N in vitro. ATPase activity was monitored at 37°C using the malachite green method in the absence or presence of a proteasome substrate, UBL-YFP-ODC (1 μg). **p<0.01, *p<0.05 (two-tailed paired Student's t-test, mean ± s.e.m from n=3 independent experiments).
(d) 26S proteasomes from 293T cells overexpressing WT or DN DYRK2 were immobilized on streptavidin beads, washed extensively with reaction buffer containing 1 mM ATP or 1 mM ATPγS, and assayed for Suc-LLVY-AMC cleavage in the same buffers. Reaction was carried out for 10 min at 37°C. **p<0.01 (two-tailed paired Student's t-test, mean ± s.e.m from n=3 independent experiments).
Source data for c and d can be found in Supplementary Table 3.