Abstract
Phytic acid (PA) biosynthesis pathway genes were reported from multiple crop species. PA accumulation was enhanced during grain filling and at that time, hormones like Abscisic acid (ABA) and Gibberellic acid (GA3) interplay to control the process of seed development. Regulation of wheat PA pathway genes has not yet been reported in seeds. In an attempt to find the clues for the regulation by hormones, the promoter region of wheat PA pathway genes was analyzed for the presence of cis-elements. Multiple cis-elements of those known to be involved for ABA, GA3, salicylic acid (SA), and cAMP sensing were identified in the promoters of PA pathway genes. Eight genes (TaIMP, TaITPK1-4, TaPLC1, TaIPK2 and TaIPK1) involved in the wheat PA biosynthesis pathway were selected for the expression studies. The temporal expression response was studied in seeds treated with ABA and GA3 using quantitative real time PCR. Our results suggested that exogenous application of ABA induces few PA pathway genes in wheat grains. Comparison of expression profiles for PA pathway for GA3 and ABA suggested the antagonistic regulation of certain genes. Additionally, to reveal stress responses of wheat PA pathway genes, expression was also studied in the presence of SA and cAMP. Results suggested SA specific differential expression of few genes, whereas, overall repression of genes was observed in cAMP treated samples. This study is an effort to understand the regulation of PA biosynthesis genes in wheat.
Keywords: phytic acid, wheat, ABA, GA3, gene regulation, seed development
1. Introduction
Gene expression is largely controlled through promoters and their contributing cis-acting elements that are positioned upstream of the transcriptional start site of a gene. These elements contain binding sites for transcription factors involved in the initiation and regulation of transcription. Specific regulation of the transcript in tissues and cells is of high significance, especially during the plant development. Most of the regulatory sequences of plant transcripts are located approximately 1000 base pairs (bp) upstream of the transcriptional start site [1,2].
Seed development and maturation is an important event that includes the process of desiccation and dormancy [3,4]. Maturation of seeds is also accompanied by gene expression responses and multiple biochemical events [5,6]. In wheat, three stages of gene expression patterns were observed during seed development. The first phase spans from three to seven days after anthesis (DAA) that includes the formation of multi-cellular tissue structure. The second stage starts from seven to fourteen DAA, which mainly involves grain filling. Lastly, seed maturation and desiccation that spans from 21 to 28 DAA [7]. The biosynthesis of several metabolic compounds are governed by hormones that mainly includes Abscisic acid (ABA) and Gibberellic acid (GA3) [8,9]. The antagonistic behaviour of ABA and GA3 plays a crucial role during seed physiology. ABA predominately regulates both accumulation of nutrients and desiccation during seed maturation, whereas GA3 activates certain enzymes required for breaking the dormancy during germination [10,11,12].
Previous findings suggested that the ABA may be a positive regulator of phytic acid (PA) synthesis in developing seeds. It has been demonstrated that ABA signal transduction is mediated by inositol-1,4,5-triphosphate (IP3) which, in turn, regulates seed maturation [13]. Studies have shown a positive correlation between ABA concentration and PA accumulation using seed suspension cells [14]. During the early phase of wheat seed development, 14 DAA aleurone was found to be present as distinct tissue from endosperm [15], and it has been reported previously that aleurone tissue is a major site for PA accumulation in most of the cereal grains [16]. In wheat, PA pathway genes are highly induced in aleurone at 14DAA. Thus, it could be highly informative to use 14DAA seeds to study the effects of exogenous hormones on PA pathway genes.
PA biosynthesis in plants utilize two pathways which are either lipid-dependent or lipid-independent [17]. The first committed step involves the formation of inositol3-phosphate (Ins3P) from glucose-6-phosphate by myo-inositol-3-phosphate synthase (MIPS). The subsequent steps involve a sequential and ordered phosphorylation of the remaining five positions of the inositol ring by a number of kinases [18,19]. The enzymes catalyzing these phosphorylation reactions include inositol monophosphatase (IMP), inositol tris/tetraphosphate kinase (ITPK), inositol polyphosphate kinase (IPK2) and inositol-pentakisphosphate 2-kinase (IPK1) [16,17,18]. Manipulating plant inositol phosphate kinases and the transporter of PA from various plant species is a key to generate low phytate crops (lpa) [18,19,20,21,22]. Additionally, IMP catalyzes the production of free inositol from dephosphorylation of Ins3P [23]. Phospholipase C (PLC) is a lipid-dependent pathway enzyme involved in the formation of IP3 utilizing phosphatidylinositol-4,5-bisphosphate (PIP2) as a substrate [18,23]. PLC activities have been implicated in processes as diverse as signal transduction including those in guard cells [24], against pathogen response [25], during gravitropism [26], Nod factor signaling [27,28]. The control of stomatal opening/closing by the plant hormone ABA represents one of the most thorough evaluations of PLC action in plants [29]. ITPK proteins have been identified for the intermediate stage of PA biosynthesis and have shown to be important in regulating Inositol tetrakisphosphate (IP4) metabolism. Multiple ITPK genes have been identified from Arabidopsis (AtITPK1-4), soybean (GmITPK1-4), rice (ITP5/6K1-6) and wheat (TaITPK1-4) [6,19,23,30]. Only one in barley (HvIPK) and maize (ZmIP5/6K) are reported [31,32]. IPK2 is a multifunctional kinase that phosphorylates IP3 at the 3rd and the 6th position. One IPK2 gene has been characterized from rice (OsIPK2) and wheat (TaIPK2) whereas, two isoforms were identified in Arabidopsis (AtIpk2α and AtIpk2β) [6,23,33]. The Inositol pentakisphosphate kinase (IPK1) is involved in the final step in phytic acid biosynthesis. This step is common to both lipid-dependent and independent pathways. One IPK1 has been characterized from rice (OsIPK1), Arabidopsis (AtIPK1) and wheat (TaIPK1) [6,23,34]. In order to understand the regulation of PA pathway genes, especially in seeds, the effect of hormones was studied previously in rice [14]. Earlier, it has been shown that the wheat MIPS gene, which is involved in earlier steps of PA biosynthesis, is inducible by ABA, salicylic acid (SA) and other abiotic stresses [35]. No additional studies regarding the hormonal regulation of the remaining wheat PA pathway genes were reported.
SA plays an important role during biotic and abiotic stress due to its ability to generate the protective response in the plants [36]. It has been known to contribute in defense signaling during seed development [37]. Recent studies suggested the role of SA in regulation of PLC in time dependent manner upon heat stress which is positively regulated by ABA treatment [38,39]. Previously, it was shown that GA3 and/or auxin influence the levels of cAMP suggesting their importance during seed maturation, germination and plant development [40,41]. Although, as suggested above, multiple roles were offered by SA and cAMP in cereal crops, but reports regarding their effect on the expression of PA pathway genes were unexplored. Therefore, it would be interesting to study the temporal expression of genes involved in phytic acid biosynthesis under exogenous application of hormones (ABA or GA3) and biochemicals (SA or cAMP). Previously, it was shown that genes responsible for the biosynthesis of PA in wheat grains were differentially regulated [6]. However, hormonal regulation of these PA pathway genes in wheat was not reported. Thus, in the current work, we performed a comprehensive expression analysis of PA pathway genes in seeds exposed to ABA, GA3, SA and cAMP. Our results suggested a coordinated and time-dependent response of the phytic acid biosynthetic genes when exogenously exposed to these hormones and signaling molecules.
2. Results
2.1. In-Silico Analysis of the Regulatory Cis-Elements
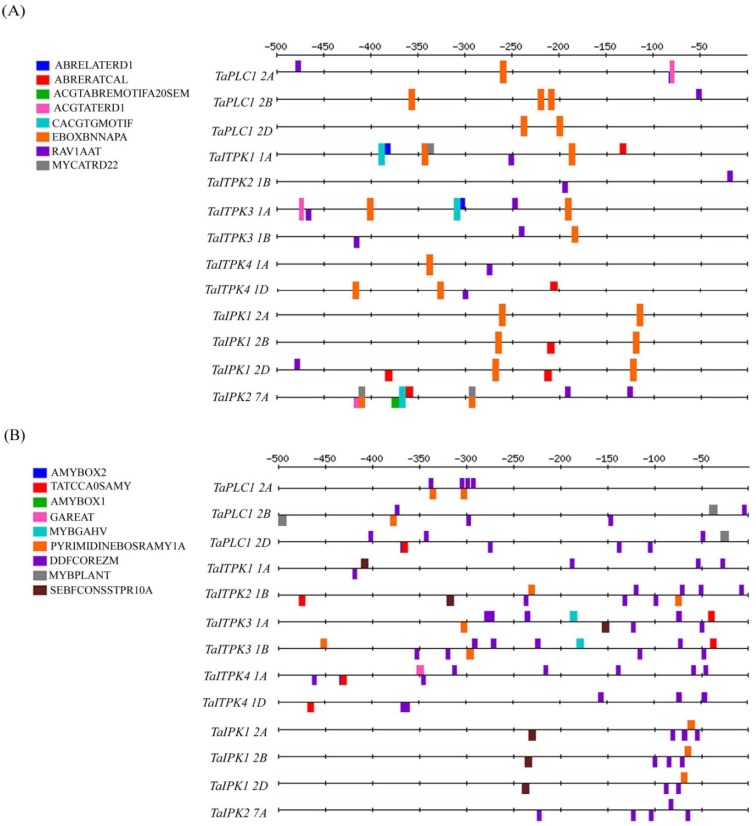
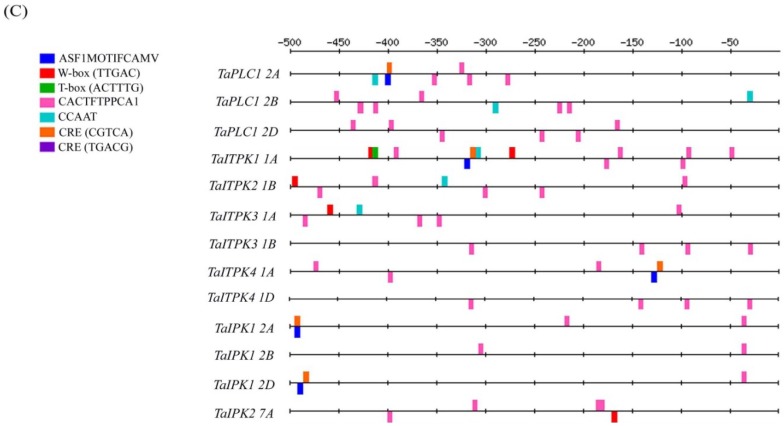
In order to elucidate the mechanism of transcriptional regulation of wheat PA pathway genes, analysis of their promoter region was performed for the cis-elements. A 1.0 kb sequence upstream to open the reading frame of TaITPK1 (Ta.70767), TaITPK2 (CA18510.1), TaITPK3 (Ta.39455), TaITPK4 (Ta.36061), TaIPK1 (Ta.41955), TaIPK2 (Ta.35113) and TaPLC1 (Ta.55212) was identified and subjected to PLACE analysis [6,42]. TaIMP was not included in promoter analysis due to its inadequate information in the International Wheat Genome Sequencing Consortium (IWGSC). Promoter analysis showed the presence of multiple cis-elements for each of these genes [43]. For the current study, we included the cis-elements of the genes those were responsive to ABA, GA3, SA and cAMP (Table A1). In general, we observed the presence of multiple numbers of responsive elements for hormones in the PA pathway genes. For more clarity, the selected cis-regulatory elements (CREs) were mapped on the basis of their location in the upstream gene region (−1 to −500 bp) using Regulatory Sequence Analysis Tools (RSAT) (Figure 1).
Figure 1.
Cis-regulatory elements (CREs) maps in the promoter (−1 to −500) of PA biosynthesis genes. The colour indicates the position of the respective cis-elements. (A) CREs map generated for ABA response elements; (B) CREs map generated for GA3response elements; (C) CREs map generated for SA and cAMP response elements.
We observed that, ABA responsive elements, EBOXBNNAPA and RAV1AAT were ubiquitously present in all of the PA pathway genes. For the remaining ABA responsive elements, either one or the other elements were present in the promoter of the homoeologous sequences. Out of all the PA pathway genes, TaPLC1 and TaIPK1 promoter elements showed highest density for the presence of ABA responsive elements. This suggests that the transcript accumulation of these genes could be easily influenced by the presence of ABA. Similarly, hormonal responsive elements for GA3 were also identified. Out of all the GA3 responsive elements, DOFCOREZM was prevalent in the promoter regions of the wheat PA pathway genes. The presence of high density of DOFCOREZM elements in all the genes suggested its importance in seeds.
In order to gather more information regarding their possible response towards signaling molecules, their promoter was also analyzed for SA and cAMP cis-elements. The known SA responsive elements that are primarily involved in repression i.e., CACTFTPPCA1 and CCAAT were present in all the genes but the highest number was observed in TaPLC1. This reinforces that the activity of PLC is regulated in the presence of stress generated signalling events. Promoter of all the wheat PA pathway genes also showed the presence of the cAMP response elements.
2.2. Hormonal Regulation of PA Pathway Genes
In order to study the temporal response of wheat PA biosynthesis pathway genes, seeds of 14 DAA were exposed to ABA and GA3 respectively. Subsequently, qRT-PCR was performed for eight genes involved in PA biosynthesis in wheat after treatment with respective hormones for 30 min, 45 min and 60 min post incubation. TaMIPS was not included in the current study since it has been characterized in detail under different abiotic stresses that include ABA, SA, NaCl and cold temperature [35]. Primers that are capable of amplifying the transcript from all the three genomes (for TaIPK1 and TaPLC1) were carefully chosen unless the gene is present as a single copy (Table A2) [6,42].
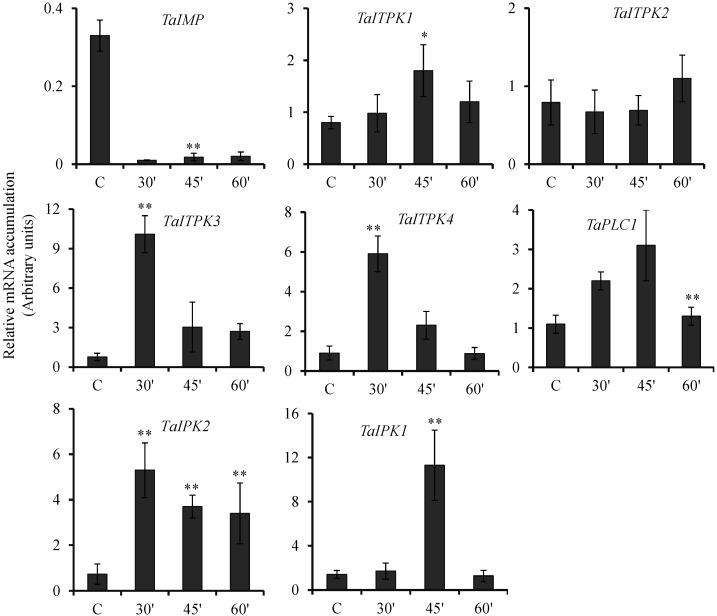
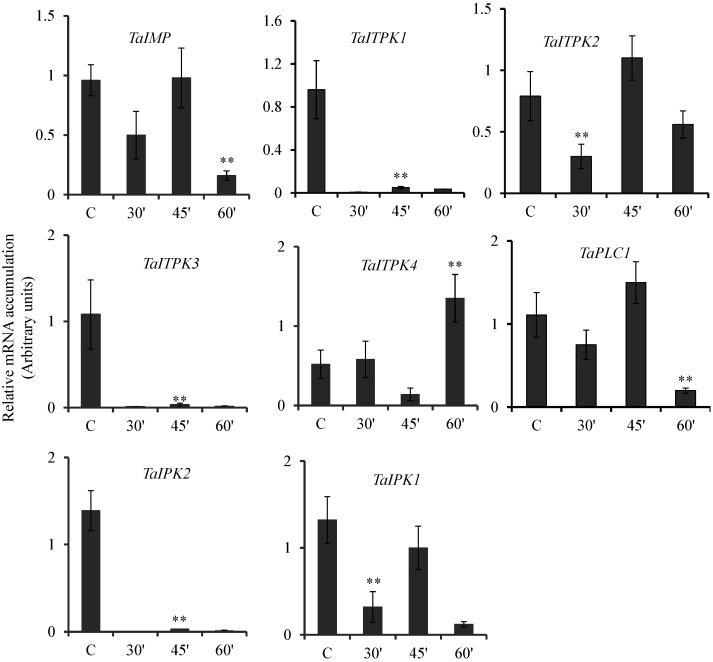
Relative fold expression levels were plotted with respect to control seeds. No significant changes in the expression of untreated samples during the time course were observed [43]. Upon exposure to ABA, wheat PA biosynthesis genes showed differential regulation during the course of the experiment (Figure 2). No significant change in the expression level of TaITPK2 was observed. TaITPK1, TaITPK3, TaITPK4, TaPLC1, TaIPK2 and TaIPK1 showed significant increase in the level of transcript at different time points. Among all the genes, the highest fold induction was observed for TaITPK3 and TaIPK1 (~10 folds) (Figure 2). Surprisingly, the expression of TaIMP was repressed as compared to control seeds. Exposure to ABA resulted in induction of six out of the eight genes studied.
Figure 2.
Effect of ABA on the expression of PA pathway genes. Relative fold expression of PA pathway genes in wheat grains exposed to ABA (100 μM) for 30, 45 and 60 min. After the exogenous application, the seeds were collected and RNA extraction was performed. The cDNA templates were prepared from 2 µg of DNase free RNA. Ct values obtained were normalized against Ta18SrRNA expression. Each bar indicates the mean of three to four replicates with the indicated standard deviation of the mean. ** indicates significant difference at p ≤ 0.01 and * indicates significant difference at p ≤ 0.05 with respect to control.
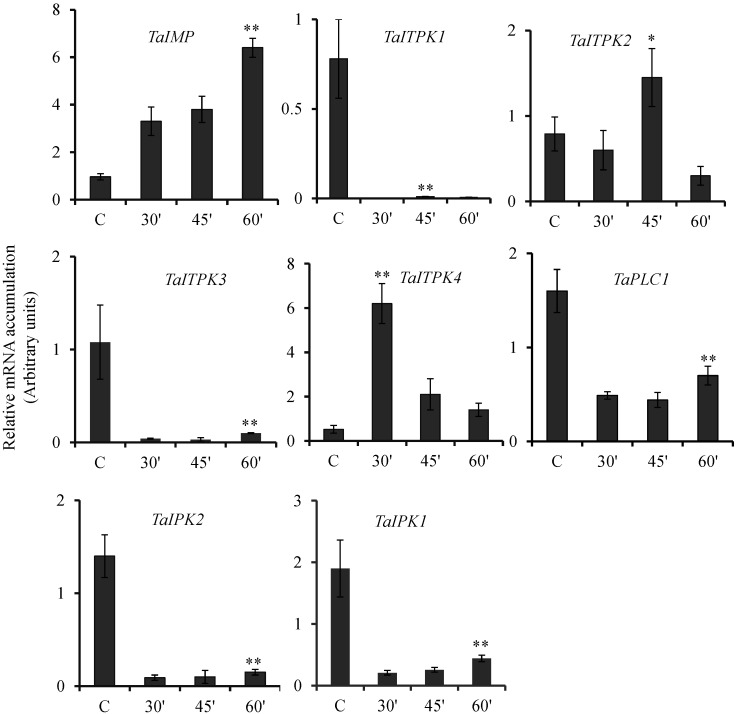
Wheat seeds exposed to GA3 showed consistently decreasing expression level for TaITPK1 and TaITPK4 (Figure 3). TaIMP was induced at the early time points studied (30 min and 45 min) and subsequently showed decreases in the transcript after 60 min post incubation. For the remaining genes, TaITPK2, TaITPK3, TaPLC1, TaIPK2 and TaIPK1 showed differential expression responses. Interestingly, expression data suggested a similar pattern of expression for TaITPK3 and TaIPK2 although at different fold levels. Likewise, the abundance of all the wheat PA biosynthesis genes except TaIMP was reduced at the last time point studied after exposure to GA3 (Figure 3). Overall, GA3 showed suppression of the five genes (TaITPK1, TaITPK3, TaITPK4, TaPLC1 and TaIPK2) that were induced by ABA, suggesting the antagonistic effects of these hormones in grains.
Figure 3.
Effect of GA3 on the expression of PA pathway genes. Relative fold expression of PA pathway genes in wheat grains exposed to GA3 (60 μM) for 30, 45 and 60 min. After the exogenous application, the seeds were collected and RNA extraction was performed. The cDNA templates were prepared from 2 µg of DNase free RNA. Ct values obtained were normalized against Ta18SrRNA expression. Each bar indicates the mean of three to four replicates with the indicated standard deviation of the mean. ** indicates significant difference at p ≤ 0.01; * indicates significant difference at p ≤ 0.05 with respect to control.
2.3. Expression Pattern of PA Pathway Genes in Presence of SA and cAMP
Multiple studies have suggested that plant resistance responses are mediated by SA [36]. Few reports also suggested that during defense activation against pathogens, PA pathway genes, especially PLC is differentially regulated. To date, no clear link of SA with the regulation of the PA pathway has been reported. Similarly, cAMP being a major signaling molecule was also included in the current study. Temporal expression of PA pathway genes was analyzed in wheat seeds exposed to SA and cAMP. Expression profile suggested significant repression in the transcript levels of TaIMP, TaITPK1, TaITPK3, TaPLC1, TaIPK2 and TaIPK1 when exposed to SA as compared to their respective controls (Figure 4). Only TaITPK4 was induced at 60 min post incubation with SA. This suggested - TaITPK4 was induced by SA in a time dependent manner. TaITPK2 showed significant decrease of the transcript at the initial time point (30 min), but this repression was recovered at 45 min. Subsequently, at 60 min, the expression returned to the basal level (Figure 4).
Figure 4.
Effect of SA on the expression of PA pathway genes. The relative expression of PA pathway genes in wheat grains exposed to SA (2 mM) for 30, 45 and 60 min. After the exogenous application, the seeds were collected and RNA extraction was performed. The cDNA templates were prepared from 2 µg of DNase free RNA. Ct values obtained were normalized against Ta18SrRNA expression. Each bar indicates the mean of three to four replicates with the indicated standard deviation of the mean. ** indicates significant difference at p ≤ 0.01; * indicates significant difference at p ≤ 0.05 with respect to control.
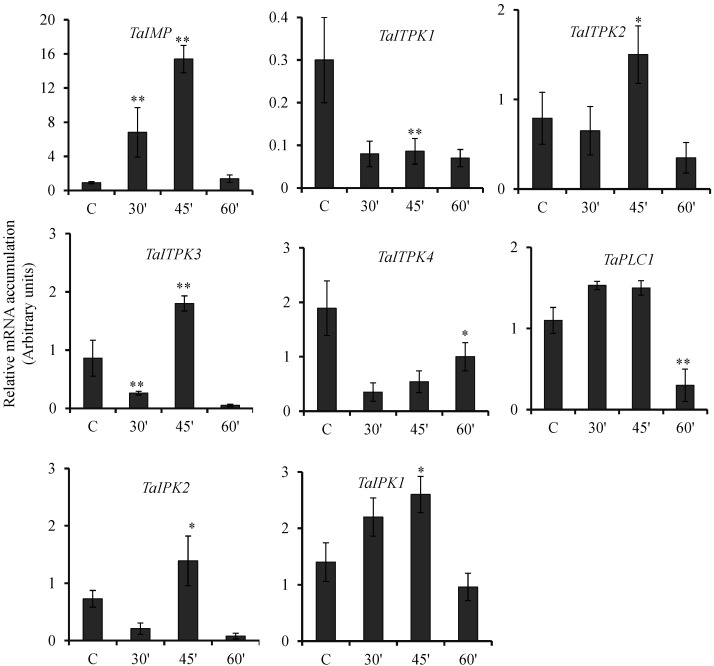
Seeds exposed to cAMP showed a concurrent increment in the expression levels of TaIMP with the time points studied. At the last time point, this increase in the level of transcript corresponds to ~5 to 6 folds as compared to the control seeds (Figure 5). TaITPK4 showed induction in their transcripts only at 30 min post incubation. Similarly, TaITPK2 showed a slight increase in the transcript levels at 45 min, although the expression reaches the basal level at 60 min. In contrast, TaITPK1, TaITPK3, TaPLC1, TaIPK2 and TaIPK1 showed significant down regulation for the respective transcripts (Figure 5). This down regulation of PA pathway genes was similar during the time course of treatment. Overall, these data suggested that wheat PA biosynthesis genes responded differently when exposed to SA or cAMP.
Figure 5.
Effect of cAMP on the expression of PA pathway genes. The relative expression of PA pathway genes in wheat grains exposed to cAMP(20 μM) for 30, 45 and 60 min. After the exogenous application, the seeds were collected and RNA extraction was performed. The cDNA templates were prepared from 2 µg of DNase free RNA. Ct values obtained were normalized against Ta18SrRNA expression. Each bar indicates the mean of three to four replicates with the indicated standard deviation of the mean. ** indicates significant difference at p ≤ 0.01; * indicates significant difference at p ≤ 0.05 with respect to control.
2.4. Expression Profile Analysis of Genes during Other Biotic and Abiotic Stresses
Multiple studies have suggested the function of plant hormones as central integrators in reprogramming developmental and stress adaptive environments [44,45,46,47,48]. To study the total effect of these biotic and abiotic stresses on the wheat PA biosynthesis pathway genes, GENEVESTIGATOR database searches were performed. The corresponding probeset IDs for each gene was retrieved from Plexdb on the basis of alignment and sequence identity (Table A3).
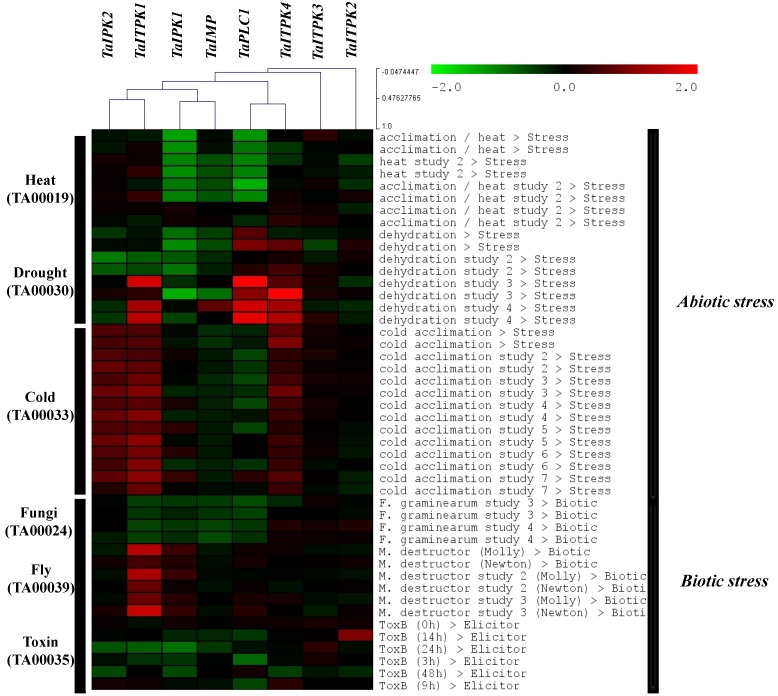
The heat map produced after an analysis revealed differential expression pattern of genes in abiotic stress and biotic stress conditions (Figure 6). Under abiotic stress conditions, TaITPK1 and TaITPK4 were preferentially induced by both cold stress and drought stress. TaIPK2 was significantly induced by cold stress, whereas TaIMP and TaPLC1 were generally downregulated in cold and heat stress environments. TaPLC1 was significantly up-regulated under drought stress. TaIPK1 was found to be down-regulated under heat stress study. To correlate the expression pattern of PA pathway genes under biotic stress conditions, studies on Fusarium graminearun (fungi), Mayetiola destructor (fly) and a fungal toxin ToxB (elicitor) were included as other biotic stress studies showed no differential expression of the PA pathway genes. The heat map showed significant up-regulation of TaITPK1 when exposed to M. destructor. Moreover, the rest of the genes showed slight downregulation under biotic stress conditions. Also, TaITPK3 showed no expression change under both abiotic and biotic stress studies. This analysis suggested that beside the prime role of PA pathway genes in seeds, they also show differential response towards multiple stresses.
Figure 6.
Microarray based expression profiles of selected PA pathway genes. Foliar expression of seven probe sets was analyzed against 250 samples representing heat, drought, cold and biotic stress conditions. The heat map was generated using meta-analysis tool at GENEVESTIGATOR (https://www.genevestigator.com/gv/plant.jsp). Colours represent the fold change for expression.
3. Discussion
Lowering the phytate content in the seeds is considered to be an important trait. Multiple reports describing the genes involved in the plant PA biosynthesis along with their functional importance have been described. Understanding the regulation of these genes remained largely unexplored. During the onset of grain maturation, the seed hormone level contributes towards the development and regulation of gene transcript [49]. Since the rate of accumulation of PA biosynthesis is higher in the early stages of seed maturation [6], we expect the effect of endogenous hormones on the expression response of PA biosynthesis genes in seeds. Thus, in the current study, we tested the effect of exogenously applied hormones (ABA and GA3) and signalling molecules (SA and cAMP) on the temporal regulation of the PA pathway genes. Initially, we identified the regulatory elements in the promoter of genes involved in the PA biosynthesis in wheat. The analysis supported our rationale as we observed multiple cis-elements in the promoter region that could be responsible for perception of either ABA, GA3, SA or cAMP (Table A1).
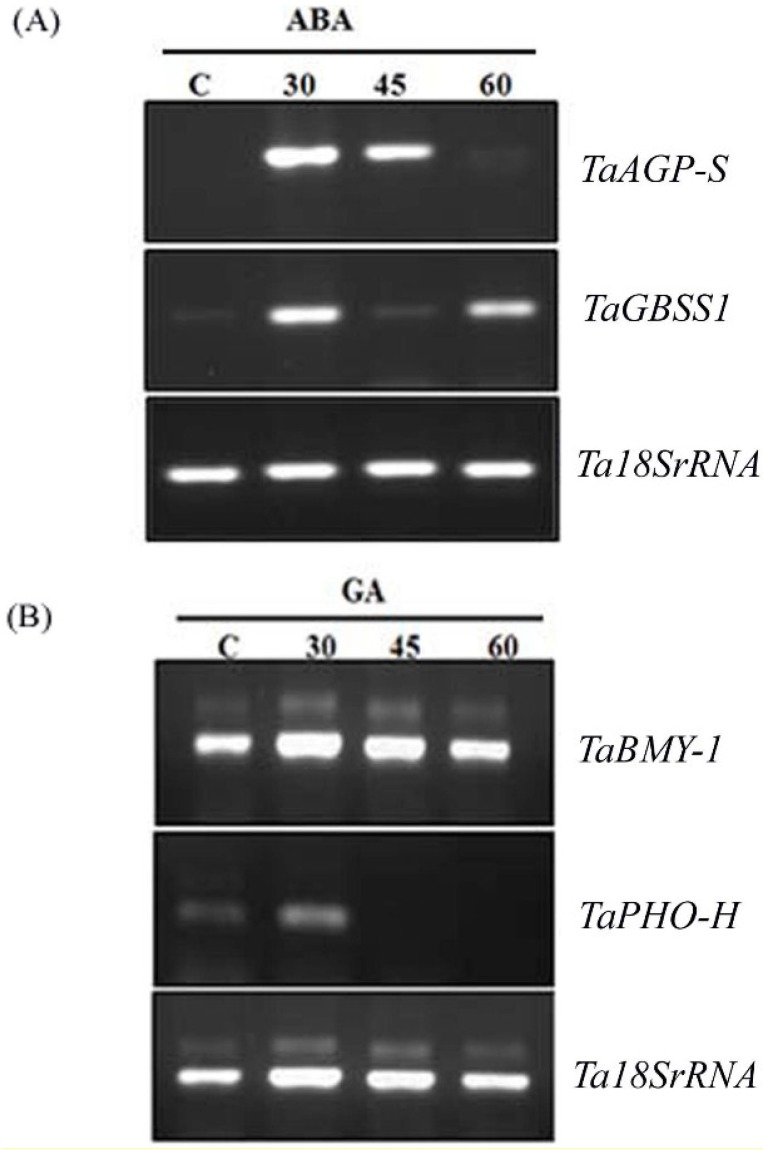
Our experimental setup (for ABA and GA3) was validated by amplifying the marker genes that included wheat amylase genes and starch synthesizing transcripts. A positive co-relation to the induction of the representative genes was observed in the respective treatment (Figure A1). Multiple copies of ABA-responsive element (RAV1AAT) and GA-responsive element (DOFCOREZM) were present in the promoters of wheat PA pathway genes suggesting their role in different physiological responses. Studies have shown that the phytic acid level is rapidly enhanced in guard cells in response to ABA [50]. Similarly, tissue specific role of phosphatidic acid signalling was also tested in barley aleurone cells [51]. These observations suggested the important role of PA pathway genes in increments of phytic acid upon exogenous ABA treatment. Enhanced PA accumulation was reported in rice suspension cells when exposed to ABA [14]. We speculate that upon ABA treatment PA accumulation could be due to the reduced breakdown of PA or its altered transport. This warrants detailed investigation for the measurement of PA in seeds treated with ABA. Interestingly, some of these cis-elements are consistently present at the same location within the promoters of the homoeologous gene. For example, in case of TaIPK1, most of the cis-elements in the promoters of different genomes were located at the similar positions.
One of the interesting observation was the earliest expression of TaITPK3 and TaITPK4 (at 30 min) followed by TaIPK1 (at 45 min) suggesting the sequential expression cascades for the PA pathway genes. In fact, one of the intermediate pathway genes in Arabidopsis, AtITPK2 was also highly induced upon ABA treatment in seeds [52]. ABA is also interlinked with the drought tolerance, for example, OsITPKgenes also showed differential response against drought stress and ABA signaling [53]. In ABA treated wheat seeds, the expression of TaIPK2 was consistently up-regulated at all time points which are in contrast to the expression of AtIPK2β that decreased upon ABA treatment [54]. Expression pattern of OsIPK1 [14], and our results regarding TaIPK1 suggested higher fold accumulation of their transcripts when treated with ABA. Except for TaITPK2 and TaIMP all the wheat genes involved in PA biosynthesis were induced by ABA (Figure 2). It will be interesting to scan the promoter region of TaIMP for the ABA-responsive cis element. This may imply regulation of the inositol signaling pathway by ABA in developing wheat seeds is pre-dominantly active downstream of TaIMP product. Treatment of wheat seeds with GA3 resulted in down-regulation of TaITPK1, TaITPK4 and TaPLC1 (Figure 3). For TaITPK3 and TaIPK2, the repression in the transcript levels was time specific. Altogether, this reinforces the observation that some of the intermediated PA pathway genes, TaITPK1, TaITPK4 and TaPLC1 are antagonistically regulated by the presence of ABA and GA3. Similarly, TaIPK1 was only induced by ABA at 45 min, whereas downregulation of the gene was observed in the presence of GA3 at 60 min. However, the expression pattern of TaIMP suggests its preferential regulation by GA3 when compared to ABA during seed development. Previously, it has been shown that GA3 leads to a rapid and transient IP3 accumulation in barley aleurone [55] and in rice seeds, ABA alone does not alter the levels of IP3 [56]. This raises a question of to what extent ABA-GA3 antagonism regulates inositol signaling leading to phytic acid. Interestingly, recent characterization of AtIMPL1 suggested it as a bi-functional enzyme and functionally different from AtIMPL2 [57,58]. AtIMPL2 does not function in the inositol signaling pathway, this functional diversification between closely related homologous enzymes warrants detailed characterization of TaIMP and its role in the inositol signaling pathway.
Overall, our studies indicated that some of the PA pathway genes are highly induced by ABA when compared to GA3. These observations are in accordance with the previous observations reported from rice suspension cells where overall induction of PA biosynthesis genes was observed in presence of ABA [14]. Altogether, our observations also support that, ABA and GA3 act antagonistically in developing wheat grains. One of the surprising observations made was the downregulation of TaIMP when exposed to ABA and SA. In contrast, two IMP in rice showed very low or no changes in their expression patterns when treated with ABA [14]. Interestingly, IMP in Cicer arietinum was induced when treated with ABA [59]. Our GENEVESTIGATOR data suggested that, under drought conditions, no significant changes in the expression TaIMP were observed. ABA has been linked to the drought conditions; thus, we anticipate that other wheat IMP may be regulated by ABA. Multiple studies have indicated the role of SA during thermo tolerance in crop plants [60]. Our studies indicated general repression of all the wheat PA pathway genes (Figure 4) in presence of SA. These observations were also reinforced by the presence of maximum number of cis-elements (mesophyll expression module1, CACTFTPPCA1 and heat shock element, CCAAT) that contributed for SA mediated repression (Table A1). These elements were over-represented in SA repressed genes [36]. On the similar lines, overall repression was observed for PA wheat genes when treated with cAMP (Figure 5). cAMP is an important signaling molecule [61], and their role in seed biology is still not explored to an extent. To get an insight of cAMP-mediated repression of PA pathway-related genes, further detailed studies need to be performed. Future studies will be directed to understanding the signaling steps that control the specific expression of PA pathway genes.
Analysis was done to correlate our expression studies to that of abiotic and biotic stresses (Figure 6). TaITPK1, TaITPK4 and TaPLC1 were induced not only by the ABA but also under drought conditions. This suggested that the response of these genes under drought conditions could be because of the ABA. Our hypothesis is supported by the in-silico observations that showed the presence of four ABA/dehydration responsive cis-elements (ACGTATERD1, ABRELATERD1, EBOXBNNAPA, MYCATRD22) [62]. Reduction of phytic acid in plants is generally accompanied with the enhanced susceptibility of the plant towards pathogens [63]. The differential response in the expression profiles of some genes (TaPLC1, TaIPK1 and TaITPK1) under biotic stresses also suggested their possible role in plant defense and in other tested conditions. Altogether, these differential response profiles for exogenous hormones, signalling molecules and other stresses suggest the importance of PA biosynthesis genes in seeds and physiology of the plants.
4. Experimental Section
4.1. Plant Materials and Growth Conditions
Bread wheat cv. C306, a good processing quality Indian variety, was used for this study. For tissue sample collection, plants were grown in growth chambers under a 12 h photoperiod at 400 μmol·m−2·s−1, 70% relative humidity and 25 °C/18 °C (day/night). To study hormonal regulation, the main individual spikes of the biological replicates were tagged at first day after anthesis (DAA). The tagged spikes were harvested at 14 DAA and were used to collect seeds.
4.2. Exogenous Treatment of Seeds with Hormones
To study the effect of hormones, abscisic acid (ABA) and gibberellic acid (GA3) were used as suggested earlier with minor modifications [51,64,65]. Six to seven spikes of 14 DAA stage were used to collect seeds and were subjected to incubation with either of the treatments that include ABA (100 μM), GA3 (60 μM), SA(2 mM) and cAMP (20 μM) at different time intervals i.e., 30, 45 and 60 min. Control seeds imbibed in CaCl2 were used. Three replicates containing 22–25 seeds per plate, with their respective hormonal treatments, were used for the experiment. Seeds were collected after the given time points and rapidly chilled before storage at −80 °C.
4.3. RNA Isolation from Plant Materials and Quantitative Real Time PCR (qRT-PCR)
RNA was extracted from wheat seeds (treated and untreated) using RNeasy Plant MiniKit (Qiagen, Valencia, CA, USA), following manufacturer instructions. Genomic DNA contamination was removed using Trubo DNA-freeTM kit(Ambion/Life Technologies, Grand Island, NY, USA). Transcriptor First Strand cDNA Synthesis Kit RT-PCR (Roche, Indianapolis, IN,USA) was used for cDNA preparation from two micrograms of RNA. Reverse transcription was performed using random hexamer primers following the manufacturer’s guidelines. Primers capable of amplying all the homeolog of the given gene were used for the study (Table A2). Thus, in the current study, an additive effect of the transcripts arising from all the different genome was represented unless the gene is present as a single copy.
Quanti-Tect SYBR Green RT-PCR Master mix (Qiagen, Valencia, CA, , USA) was used in qRT-PCR reactions,upto 45 cycles on 7500 Fast & 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Five replicates from each biological sample were used to perform qRT-PCR analysis. For PCR reaction, four replicates for each gene were amplified from two independent cDNA preparations. Relative expression level was quantified using 2−ΔΔCt method after normalizing Ct values against 18 s rRNA expression [66,67].
4.4. In-Silico Analysis of Cis-Element Search in Upstream Region of Wheat Genes
The full length cDNA sequence of respective genes was confirmed using NCBI (Blastx). The promoter sequences were obtained by comparison with wheat genomic sequences. One-thousand bp upstream of 5'-terminus of each cDNA were selected by using genomic data from IWGSC (http://wheat-urgi.versailles.inra.fr/Seq-Repository/). Due to limited genomic sequence information about wheat genome, promoter sequences for TaITPK3, TaITPK4 and TaIPK2 were derived by using Triticum Urartu (A-genome) or Aegilops tauschii (D-genome) genomic database from NCBI or IWGSC. Table A4 describes the sequence IDs for each gene that were used to extract promoter regions. Promoter sequence for TaIMP could not be retrieved due to inadequate information in IWGSC. All possible cis-elements known in plants were searched by using PLACE database (http://www.dna.affrc.go.jp/PLACE/). The cis-regulatory elements (CREs) were mapped −1 to −500 bp upstream gene region using RSAT tool (http://www.rsat.eu/).
4.5. GENEVESTIGATOR Analysis of PA Pathway Genes
The spatial distribution of genes involved in phytic acid synthesis was studied using seven probe set IDs (Table A3). The expression pattern of PA pathway genes was analyzed across 250 samples representing heat, drought, cold and biotic stress conditions. Microarray expression IDs from PlexDB (TA00019, TA00030, TA00033, TA00024, TA00039 and TA00035) representing the above stresses was used for generating heat maps. They were clustered into heat maps to compare expression of candidate genes using GENEVESTIGATOR software [68]. In addition, Euclidean distance matrix algorithms using optimal leaf-ordering was used to hierarchically cluster the expression data.
5. Conclusions
In conclusion, we have observed that ABA primarily controls the expression of wheat PA pathway genes. These genes are also regulated under multiple stresses and by other signalling molecules. Such hormone-based regulation studies of multiple genes involved in biological pathways could be important for genetic interventions aimed at trait development. Additionally, our expression profiles for the wheat PA pathway genes suggested the antagonistic relation of ABA and GA3 in seeds.
Acknowledgments
The authors would like to thank Executive Director, NABI for facilities and support. Part of this research was supported by a Department of Biotechnology (Grant No: BT/PR5989/AGII/106/867/2012) awarded to Ajay Kumar Pandey. Sipla Aggarwal was supported by Indian Council of Medical Research, Govt. of India for fellowships. Kaushal Kumar Bhati acknowledges Department of Biotechnology , Mandeep Kaur acknowledges Council of Scientific and Industrial Research-University Grant Commission, Govt. of India, for fellowships.
Appendix
Figure A1.
Semi-quantitative RT-PCR of wheat genes as a marker for ABA and GA3 treatment on seeds. (A) PCR amplification of TaAGP-S (ADP-glucose pyrophosphorylase, smaller subunit) and TaGBSS1 (Granule-bound starch synthase 1) from 14 DAA seeds treated with ABA; (B) PCR amplification of TaBYM1 (β-amylase) and TaPHO-H (starch phosphorylase H) in wheat seeds (14 DAA) when treated with GA3. After respective treatments the seeds were collected at 30 min, 45 min and 60 min post incubation. 2 µg of total RNA (DNase treated)was extracted and cDNA templates were prepared. Low-cycle semi-quantitative RT-PCR was performed along with Ta18SrRNA as an internal control. The amplicons were resolved on 1.2% agarose gel. The experiments were repeated twice from different biological replicates.
Table A1.
Specitication of cis-responsive elements of ABA, GA3, SA and cAMP in genes involved during PA biosynthesis in wheat. The number below represents the occourance of the response elements in the promoter.
| cis elements | TaPLC1 | TaITPK1 | TaITPK2 | TaITPK3 | TaITPK4 | TaIPK2 | TaIPK1 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2A | 2B | 2D | 1A | 1B | 1A* | 1B | 1A* | 1D* | 7A* | 2A | 2B | 2D | ||
| ABA RESPONSE ELEMENTS | ||||||||||||||
| ABRELATERD1 | 1 | 0 | 1 | 3 | 0 | 3 | 0 | 1 | 1 | 4 | 1 | 2 | 1 | |
| ABRERATCAL | 1 | 0 | 1 | 2 | 1 | 1 | 0 | 0 | 1 | 2 | 2 | 4 | 4 | |
| ACGTABREMOTIFA2OSEM | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |
| ACGTATERD1 | 4 | 4 | 4 | 6 | 0 | 6 | 0 | 2 | 4 | 8 | 2 | 4 | 2 | |
| CACGTGMOTIF | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | |
| EBOXBNNAPA | 6 | 10 | 8 | 8 | 14 | 10 | 10 | 8 | 12 | 6 | 10 | 10 | 12 | |
| RAV1AAT | 5 | 8 | 4 | 2 | 3 | 3 | 3 | 2 | 2 | 4 | 2 | 1 | 2 | |
| MYCATRD22 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 2 | 1 | 1 | 1 | |
| GA RESPONSE ELEMENTS | ||||||||||||||
| AMYBOX2 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |
| TATCCAOSAMY | 0 | 0 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | |
| AMYBOX1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 2 | |
| GAREAT | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 2 | |
| MYBGAHV | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 2 | |
| PYRIMIDINEBOXOSRAMY1A | 3 | 1 | 0 | 0 | 2 | 1 | 2 | 0 | 0 | 0 | 2 | 3 | 2 | |
| DOFCOREZM | 9 | 7 | 9 | 6 | 10 | 17 | 12 | 13 | 17 | 7 | 7 | 9 | 9 | |
| MYBPLANT | 0 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | |
| SEBFCONSSTPR10A | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 2 | 3 | 2 | 2 | |
| SA RESPONSE ELEMENTS | ||||||||||||||
| ASF1MOTIFCAMV | 4 | 1 | 0 | 1 | 1 | 0 | 1 | 4 | 2 | 2 | 1 | 0 | 2 | |
| W-box (TTGAC) | 0 | 1 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 5 | 0 | 1 | 1 | |
| T-box (ACTTTG) | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | |
| CACTFTPPCA1 | 10 | 12 | 15 | 14 | 12 | 12 | 11 | 13 | 10 | 10 | 8 | 10 | 8 | |
| CCAAT | 2 | 2 | 2 | 4 | 1 | 1 | 1 | 1 | 2 | 1 | 0 | 0 | 0 | |
| cAMP RESPONSE ELEMENTS# | ||||||||||||||
| CRE (CGTCA) | 2 | 1 | 0 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | |
| CRE (TGACG) | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 1 | |
* Promoter gene sequence derived from Triticum urartu (A-genome) or Aegilops tauschii (D-genome); # CRE elements consensus sequences from were manually searched in the promoter sequences [69].
Table A2.
Primer sequences used in the current study.
| Name of Gene | Primers (5'–3') | Amplicon size (in bp) |
|---|---|---|
| TaITPK1 | Forward: ATGGTGGCCGAGCACCAGTCCTC Reverse: GACGACGGGCACGGAGGGGTGG | 261 |
| TaITPK2 | Forward: AGGAGTTCGTCAACCATGGCGGCGT Reverse: GCCGCCCGCGATCTGGTTGATGAAT | 247 |
| TaITPK3 | Forward: ACGCCATCGACCGCCTCCACAACC Reverse: CGGCGACGAGGGGCTTGGCGATGA | 222 |
| TaITPK4 | Forward: CAGCCTGCCGGACGTGCCGAC Reverse: ATGTCGAAGTTGAACAGCTGCA | 210 |
| TaPLCl | Forward: CGTGCTCCTATCAACAAAGCC Reverse : CTGTTCGTCCTCATCGTCGT | 317 |
| TaITPK2 | Forward: GCCCTACGTCACCAAGTGCCTCGC Reverse: AACCACGCCTTGAGCTCGCGCAGC | 283 |
| TaITPK1 | Forward: ACTGGGTCTACAAGGGAGAGGGC Reverse : ACACGAACCCCGCCATCAACATGA | 279 |
| TaLMP | Forward: GGACCACAAGTTCATCGGCGAGG Reverse: CTCCAACGGTGGGAATCTTCCC | 176 |
| Ta18SrRNA | Forward: GTGACGGGTGACGGAGAATT Reverse : GACACTAATGCGCCCGGTAT | 150 |
| TaAGP- S | Forward: GCAAGATACACCATTCAGTAGTTGGAC Reverse : GACTGTTCCACTAGGGAGTAAAGCATC | 326 |
| TaGBSS-I | Forward: CTCGCCGCCAACTACGACGTC Reverse : TGCTCGGGAACTTCTCCTCCAC | 268 |
| TaBMY | Forward: CTAGCCAACTATGTCCAAGTCTACGT Reverse : ACTGTGGGATGGGGATGTTGACGA | 370 |
| TaSP-H | Forward: TGGCCAGCAAAAAGCGCCTAGC Reverse : TCTGCCTGTCTGCTGCGCTCA | 183 |
Table A3.
Alignment table for gene vs. probesets. PA pathway gene sequences were queried by Blastn against Plexdb wheat microarray database.
| Query gene | Target Probe set ID | e-value | Identity (%) | Query length | Query range | Target length | Target range | Target description |
|---|---|---|---|---|---|---|---|---|
| TaITPK1 | Ta.27561.1.S1_at | 0.0 | 97.2 | 1038 | 25..938 | 1219 | 120..1036 | gb|CK215630 |
| TaITPK2 | TaAffx.113052.1.S1_at | 9e-52 | 86.2 | 876 | 533..770 | 549 | 139..377 | gb|CA618510 |
| TaITPK3 | TaAffx.19998.1.A1_at | 0.0 | 97.5 | 1057 | 490..1057 | 1696 | 616..1184 | gb|CD867474 |
| TaITPK3 | TaAffx.19998.1.A1_at | 1e-151 | 97.0 | 1057 | 134..431 | 1696 | 1243..1540 | gb|CD867474 |
| TaITPK3 | TaAffx.19998.1.A1_at | 1e-29 | 100.0 | 1057 | 49..114 | 1696 | 1560..1625 | gb|CD867474 |
| TaITPK4 | TaAffx.121326.1.S1_at | 0.0 | 99.0 | 1018 | 6..606 | 722 | 64..667 | gb|CA690518 |
| TaITPK4 | Ta.27561.1.S1_x_at | 3e-58 | 84.4 | 1018 | 54..342 | 1219 | 134..422 | gb|CK215630 |
| TaPLC1 | Ta.13185.2.A1_at | 1e-46 | 86.5 | 1876 | 1541..1750 | 224 | 1..204 | gb|BJ214989 |
| TaIPK2 | Ta.30464.1.A1_at | 1e-133 | 100.0 | 899 | 1-240 | 858 | 291-530 | gb|CK214494 |
| TaIPK2 | Ta.30464.1.A1_at | 1e-117 | 100.0 | 899 | 306-517 | 858 | 14-225 | gb|CK214494 |
| TaIPK1 | TaAffx.123037.2.S1_at | 0.0 | 96.4 | 1125 | 28-1125 | 1502 | 198-1297 | gb|CA613702 |
| TaIMP | Ta.14044.1.S1_at | 0.0 | 97.5 | 983 | 1..893 | 1085 | 90..982 | gb|BT009424 |
Table A4.
Sequence IDs for each gene those were used to extract promoter regions.
| Query gene | IWGSC ID | NCBI ID |
|---|---|---|
| TaITPK1-2A | IWGSC_1AL_v2|IWGSC_chr1AL_v2_ab_k71_contigs_longerthan_200_3977071 | |
| TaITPK21B | IWGSC_1BL|IWGSC_chr1BL_ab_k71_contigs_longerthan_200_3850458 | |
| TaITPK3-1A* | - | gb|KD008473.1 |
| TaITPK3-1B | IWGSC_1BL|IWGSC_chr1BL_ab_k71_contigs_longerthan_200_5693968 | |
| TaITPK4-1A* | - | gb|KD262384.1 |
| TaITPK4-1D* | - | gb|KD502010.1 |
| TaPLC1-2A | IWGSC_2AL|IWGSC_chr2AL_ab_k71_contigs_longerthan_200_6422019 | |
| TaPLC1-2B | IWGSC_2BL|IWGSC_chr2BL_ab_k71_contigs_longerthan_200_7936983 | |
| TaPLC1-2D | IWGSC_2DL|IWGSC_chr2DL_ab_k71_contigs_longerthan_200_9907826 | |
| TaIPK2-7A* | - | gb|KD115052.1 |
| TaIPK1-2A | IWGSC_2AL|IWGSC_chr2AL_ab_k71_contigs_longerthan_200_6422019 | |
| TaIPK1-2B | IWGSC_2BL|IWGSC_chr2BL_ab_k71_contigs_longerthan_200_7936983 | |
| TaIPK1-2D | IWGSC_2DL|IWGSC_chr2DL_ab_k71_contigs_longerthan_200_9907826 |
Author Contributions
Sipla Aggarwal, Ajay Kumar Pandey and Shrikant Mantri conceived and designed the experiments; Sipla Aggarwal and Vishnu Shukla performed the experiments; Sipla Aggarwal, Ajay Kumar Pandey and Kaushal Kumar Bhati analyzed the data; Sipla Aggarwal, Vishnu Shukla, Mandeep Kaur, Shivani Sharma and Anuradha Singh contributed reagents/materials/analysis tools; Ajay Kumar Pandey, Sipla Aggarwal, Vishnu Shukla and Shrikant Mantri wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Dean C., Schmidt R. Plant genomes a current description. Plant Physiol. Plant Mol. Biol. 1995;46:395–418. doi: 10.1146/annurev.pp.46.060195.002143. [DOI] [Google Scholar]
- 2.Yamamoto Y.Y., Ichida H., Matsui M., Obokata J., Sakurai T., Satou M., Seki M., Shinozaki K., Abe T. Identification of plant promoter constituents by analysis of local distribution of short sequences. BMC Genomics. 2007;8:1471–2164. doi: 10.1186/1471-2164-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bewley J.D. Seed germination and dormancy. Plant Cell. 1997;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Domínguez F., Cejudo F.J. Programmed cell death (PCD): An essential process of cereal seed development and germination. Front. Plant Sci. 2014;5:366. doi: 10.3389/fpls.2014.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams S.G. The Role of Phytic Acid in the Wheat Grain. Plant Physiol. 1970;45:376–381. doi: 10.1104/pp.45.4.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhati K.K., Aggarwal S., Sharma S., Mantri S., Singh S.P., Bhalla S., Kaur J., Tiwari S., Roy J.K., Tuli R., et al. Differential expression of structural genes for the late phase of phytic acid biosynthesis in developing seeds of wheat (Triticumaestivum L.) Plant Sci. 2014;224:74–85. doi: 10.1016/j.plantsci.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Laudencia-Chingcuanco D.L., Stamova B.S., You F.M., Lazo G.R., Beckles D.M., Anderson O.D. Transcriptional profiling of wheat caryopsis development using cDNA microarrays. Plant Mol. Biol. 2007;63:651–668. doi: 10.1007/s11103-006-9114-y. [DOI] [PubMed] [Google Scholar]
- 8.Fincher G.B. Molecular and cellular biology associated with endosperm mobilization in germinating cereal grains. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1989;40:305–346. doi: 10.1146/annurev.pp.40.060189.001513. [DOI] [Google Scholar]
- 9.Jones R.L., Jacobsen J.V. Regulation of the synthesis and transport of secreted proteins in cereal aleurone. Int. Rev. Cytol. 1991;126:49–88. doi: 10.1016/s0074-7696(08)60682-8. [DOI] [PubMed] [Google Scholar]
- 10.White C.N., Proebsting W.M., Hedden P., Rivin C.J. Gibberellins and Seed Development in Maize. I. Evidence That Gibberellin/Abscisic Acid Balance Governs Germination versus Maturation Pathways. Plant Physiol. 2000;122:1081–1088. doi: 10.1104/pp.122.4.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bethke P.C., Hwang Y.S., Zhu T., Jones R.L. Global patterns of gene expression in the aleurone of wild-type and dwarf1 mutant rice. Plant Physiol. 2006;140:484–498. doi: 10.1104/pp.105.074435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y., Ye N., Liu R., Chen M., Zhang J. H2O2 mediates the regulation of ABA catabolism and GA biosynthesis in Arabidopsis seed dormancy and germination. J. Exp. Bot. 2010;61:2979–2990. doi: 10.1093/jxb/erq125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taji T., Takahashi S., Shinozaki K. Inositols and their metabolites in abiotic and biotic stress responses. Subcell. Biochem. 2006;39:239–264. doi: 10.1007/0-387-27600-9_10. [DOI] [PubMed] [Google Scholar]
- 14.Matsuno K., Fujimura T. Induction of phytic acid synthesis by abscisic acid in suspension-cultured cells of rice. Plant Sci. 2014;217–218:152–157. doi: 10.1016/j.plantsci.2013.12.015. [DOI] [PubMed] [Google Scholar]
- 15.Gillies S.A., Futardo A., Henry R.J. Gene expression in the developing aleurone and starchy endosperm of wheat. Plant Biotechnol. J. 2012;10:668–679. doi: 10.1111/j.1467-7652.2012.00705.x. [DOI] [PubMed] [Google Scholar]
- 16.Raboy V. Approaches and challenges to engineering seed phytate and total phosphorus. Plant Sci. 2009;79:281–296. doi: 10.1016/j.plantsci.2009.06.012. [DOI] [Google Scholar]
- 17.Stevenson-Paulik J., Bastidas R.J., Chiou S.T., Frye R.A., York J.D. Generation of phytate free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proc. Natl. Acad. Sci. USA. 2005;102:12612–12617. doi: 10.1073/pnas.0504172102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasmussen S.K., Ingvardsen C.R., Torp A.M. Mutations in genes controlling the biosynthesis and accumulation of inositol phosphates in seeds. Biochem. Soc. Trans. 2010;38:689–694. doi: 10.1042/BST0380689. [DOI] [PubMed] [Google Scholar]
- 19.Stiles A.R., Qian X., Shears S.B., Grabau E.A. Metabolic and signaling properties of an Itpk gene family in Glycine max. FEBS Lett. 2008;582:1853–1858. doi: 10.1016/j.febslet.2008.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panzeri D., Cassani E., Doria E., Tagliabue G., Forti L., Campion B., Bollini R., Brearley C.A., Pilu R., Nielsen E., et al. A defective ABC transporter of the MRP family, responsible for the bean lpa1 mutation, affects the regulation of the phytic acid pathway, reduces seed myo-inositol and alters ABA sensitivity. New Phytol. 2011;191:70–83. doi: 10.1111/j.1469-8137.2011.03666.x. [DOI] [PubMed] [Google Scholar]
- 21.Raboy V. Seeds for a better future: “Low phytate” grains help to overcome malnutrition and reduce pollution. Trends Plant Sci. 2001;6:458–462. doi: 10.1016/S1360-1385(01)02104-5. [DOI] [PubMed] [Google Scholar]
- 22.Raboy V. The ABCs of low-phytate crops. Nat Biotechnol. 2007;25:874–875. doi: 10.1038/nbt0807-874. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki M., Tanaka K., Kuwano M., Yoshida K.T. Expression pattern of inositolphosphate-related enzymes in rice (Oryzasativa L.): Implications for the phytic acid biosynthetic pathway. Gene. 2007;405:55–64. doi: 10.1016/j.gene.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Hunt L., Leckie C.P., Aitken F.L., Wentworth M., McAinsh M.R., Gray J.E., Hetherington A.M. The effects of manipulating phospholipase C on guard cell ABA-signalling. J. Exp. Biol. 2004;55:199–204. doi: 10.1093/jxb/erh027. [DOI] [PubMed] [Google Scholar]
- 25.De-Jong C.F., Laxalt A.M., Bargmann B.O., de Wit P.J., Joosten M.H., Munnik T. Phosphatidic acid accumulation is an early response in the Cf-4/Avr4 interaction. Plant J. 2004;39:1–12. doi: 10.1111/j.1365-313X.2004.02110.x. [DOI] [PubMed] [Google Scholar]
- 26.Perera I.Y., Heilmann I., Chang S.C., Boss W.F., Kaufman P.B. A role for inositol 1,4,5-trisphosphate in gravitropicsignaling and the retention of cold-perceived gravistimulation of oat shoot pulvini. Plant Physiol. 2001;125:1499–1507. doi: 10.1104/pp.125.3.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charron D., Pingret J.L., Chabaud M., Journet E.P., Barker D.G. Pharmacological evidence that multiple phospholipid signaling pathways link rhizobium nodulation factor perception in Medicagotruncatula root hairs to intracellular responses, including Ca2+ spiking and specific ENOD gene expression. Plant Physiol. 2004;136:3582–3593. doi: 10.1104/pp.104.051110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hartog M.D., Verhoef N., Munnik T. Nod factors and elicitors activate different phospholipid signalling pathways in suspension cultured alfalfa cells. Plant Physiol. 2003;132:311–317. doi: 10.1104/pp.102.017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacob T., Ritchie S., Assmann S.M., Gilroy S. Abscisic acid signal transduction in guard cells is mediated by phospholipase D activity. Proc. Natl. Acad. Sci. USA. 1999;9:12192–12197. doi: 10.1073/pnas.96.21.12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sweetman D., Stavridou I., Johnson S., Green P., Caddick S.E., Brearley C.A. Arabidopsis thaliana inositol 1,3,4-trisphosphate 5/6-kinase 4 (AtITPK4) is an outlier to a family of ATP-grasp fold proteins from Arabidopsis. FEBS Lett. 2007;581:4165–4417. doi: 10.1016/j.febslet.2007.07.046. [DOI] [PubMed] [Google Scholar]
- 31.Josefsen L., Bohn L., Sørensen M.B., Rasmussen S.K. Characterization of a multifunctional inositol phosphate kinase from rice and barley belonging to the ATP-grasp superfamily. Gene. 2007;397:114–125. doi: 10.1016/j.gene.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 32.Shi J., Wang H., Wu Y., Hazebroek J., Meeley R.B., Ertl D.S. The maize low-phytic acid mutant lpa2 is caused by mutation in an inositol phosphate kinase gene. Plant Physiol. 2003;131:507–515. doi: 10.1104/pp.014258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stevenson-Paulik J., Odom A.R., York J.D. Molecular and biochemical characterization of two plant inositol polyphosphate 6-/3-/5-kinases. J. Biol. Chem. 2002;277:42711–42718. doi: 10.1074/jbc.M209112200. [DOI] [PubMed] [Google Scholar]
- 34.Sweetman D., Johnson S., Caddick S.E., Hanke D.E., Brearley C.A. Characterization of an Arabidopsis inositol 1,3,4,5,6-pentakisphosphate 2-kinase (AtIPK1) Biochem. J. 2006;394:95–103. doi: 10.1042/BJ20051331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khurana N., Chauhan H., Khurana P. Expression analysis of a heat-inducible, Myo-inositol-1-phosphate synthase (MIPS) gene from wheat and the alternatively spliced variants of rice and Arabidopsis. Plant Cell Rep. 2012;31:237–325. doi: 10.1007/s00299-011-1160-5. [DOI] [PubMed] [Google Scholar]
- 36.Ruelland E., Pokotylo I., Djafi N., Cantrel C., Repellin A., Zachowski A. Salicylic acid modulates levels of phosphoinositide dependent-phospholipase C substrates and products to remodel the Arabidopsis suspension cell transcriptome. Front. Plant Sci. 2014;5:608. doi: 10.3389/fpls.2014.00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burkart-Waco D., Ngo K., Dilkes B., Josefsson C., Comai L. Early disruption of maternal-zygotic interaction and activation of defense-like responses in arabidopsis interspecific crosses. Plant Cell. 2013;6:2037–2055. doi: 10.1105/tpc.112.108258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu H.-T., Liu Y.-Y., Pan Q.-H., Yang H.-R., Zhan J.-C., Huang W.-D. Novel interrelationship between salicylic acid, abscisic acid, and PIP2-specific phospholipase C in heat acclimation-induced thermotolerance in pea leaves. J. Exp. Bot. 2006;12:3337–3347. doi: 10.1093/jxb/erl098. [DOI] [PubMed] [Google Scholar]
- 39.Liu H.-T., Huang W.-D., Pan Q.-H., Weng F.-H., Zhan J.-C., Liu Y., Wan S.-B., Liu Y.-Y. Contributions of PIP2-specificphospholipase C and free salicylic acid to heat acclimation-induced thermotolerance in pea leaves. J. Plant Physiol. 2006;163:405–416. doi: 10.1016/j.jplph.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 40.Pollard C.J. Influence of gibbrellic acid on the incorporation of 8–14C adenine adenosine 3',5'-cyclic phosphate in barley aleurone layers. Biochim. Biophys. 1970;201:511–512. doi: 10.1016/0304-4165(70)90176-5. [DOI] [PubMed] [Google Scholar]
- 41.Moutinho A., Hussey P.J., Trewavas A.J., Malho R. cAMP acts as a second messenger in pollen tube growth and reorientation. Proc. Natl. Acad. Sci. USA. 2001;18:10481–10486. doi: 10.1073/pnas.171104598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang K., Jin C., Wu L., Hou M., Dou S. Expression analysis of a stress-related phosphoinositide-specific phospholipase C gene in wheat (Triticum aestivum L.) PLoS ONE. 2014;9:e105061. doi: 10.1371/journal.pone.0105061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aggarwal S., Shukla V., Mantri S., Pandey A.K. National Agri-Food Biotechnology Institute; Mohali, India: 2015. Unpublished data. [Google Scholar]
- 44.Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2009;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 45.Raghavendra A.S., Gonugunta V.K., Christmann A., Grill E. ABA perception and signalling. Trends Plant Sci. 2010;15:395–401. doi: 10.1016/j.tplants.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 46.Weiner J.J., Peterson F.C., Volkman B.F., Cutler S.R. Structural and functional insights into core ABA signaling. Curr. Opin. Plant Biol. 2010;13:495–502. doi: 10.1016/j.pbi.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh B., Usha K. Salicylic acid induced physiological and biochemical changes in wheat seedlings under water stress. Plant Growth Regul. 2003;39:137–141. doi: 10.1023/A:1022556103536. [DOI] [Google Scholar]
- 48.Shakirova F.M., Sakhabutdinova A.R., Bezrukova M.V., Fathutdinova R.A., Fathutdinova D.R. Changes in hormonal status of wheat seedlings induced by salicylic acid and salinity. Plant Sci. 2003;164:317–322. doi: 10.1016/S0168-9452(02)00415-6. [DOI] [Google Scholar]
- 49.Kong L., Guo H., Sun M. Signal transduction during wheat grain development. Planta. 2015;241:789–801. doi: 10.1007/s00425-015-2260-1. [DOI] [PubMed] [Google Scholar]
- 50.Finkelstein R.R., Gampala S.S., Rock C.D. Abscisic acid signaling in seeds andseedlings. Plant Cell. 2002;14:S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Villasuso A.L., Di-Palma M.A., Pasquaré S.J., Racagni G., Giusto N.M., Machado E.E. Differences in phosphatidic acid signalling and metabolism between ABA and GA treatments of barley aleuronecells. Plant Physiol. Biochem. 2013;65:1–8. doi: 10.1016/j.plaphy.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 52.Tang Y., Tan S., Xue H. Arabidopsis inositol 1,3,4-trisphosphate 5/6 kinase 2 is required for seed coat development. Acta Biochim. Biophys. Sin. 2013;45:549–560. doi: 10.1093/abbs/gmt039. [DOI] [PubMed] [Google Scholar]
- 53.Du H., Liu L., You L., Yang M., He Y., Li X., Xiong L. Characterization of an inositol 1,3,4-trisphosphate 5/6-kinase gene that is essential for drought and salt stress responses in rice. Plant Mol. Biol. 2011;77:547–563. doi: 10.1007/s11103-011-9830-9. [DOI] [PubMed] [Google Scholar]
- 54.Yang L., Tang R., Zhu J., Liu H., Mueller-Roeber B., Xia H., Zhang H. Enhancement of stress tolerance in transgenic tobacco plants constitutively expressing AtIpk2β, an inositol polyphosphate 6-/3-kinase from Arabidopsis thaliana. Plant Mol. Biol. 2008;66:329–343. doi: 10.1007/s11103-007-9267-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Villasuso A.L., Molas M.L., Racagni G., Abdala G., Machado-Domenech E. Gibberellin signal in barley aleurone: Early activation of PLC by G protein mediates amylase secretion. Plant Growth Regul. 2003;41:197–205. doi: 10.1023/B:GROW.0000007505.37356.89. [DOI] [Google Scholar]
- 56.Kashem M.A., Itoh K., Iwabuchi S., Hori H., Mitsui T. Possible involvement of phosphoinositide-Ca2+ signaling in the regulation of α-amylase expression and germination of rice seed (Oryza sativa L.) Plant Cell Physiol. 2000;41:399–407. doi: 10.1093/pcp/41.4.399. [DOI] [PubMed] [Google Scholar]
- 57.Nourbakhsh A., Collakova E., Gillaspy G.E. Characterization of the inositol monophosphatase gene family in Arabidopsis. Front. Plant Sci. 2014;5:725. doi: 10.3389/fpls.2014.00725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petersen L.N., Marineo S., Mandala S., Davids F., Sewell B.T., Ingle R.A. The missing link in plant histidine biosynthesis: Arabidopsis myo-inositol monophosphatase-like2 encodes a functional histidinol-phosphate phosphatase. Plant Physiol. 2010;152:1186–1196. doi: 10.1104/pp.109.150805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saxena S.C., Salvi P., Kaur H., Verma P., Petla B.P., Rao V., Kamble N., Majee M. Differentially expressed myo-inositol monophosphatase gene (CaIMP) in chickpea (Cicer arietinum L.) encodes a lithium-sensitive phosphatase enzyme with broad substrate specificity and improves seed germination and seedling growth under abiotic stresses. J. Exp. Bot. 2013;64:5623–5639. doi: 10.1093/jxb/ert336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dat J.F., Lopez-Delgado H., Foyer C.H., Scott I.M. Parallel changes in H2O2 and catalase during thermotolerance induced by salicylic acid or heat acclimation in mustard seedlings. Plant Physiol. 1998;116:1351–1357. doi: 10.1104/pp.116.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gehring C. Adenylcyclases and cAMP in plant signaling-past and present. Cell Commun. Signal. 2010;8 doi: 10.1186/1478-811X-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yazaki J., Shimatani Z., Hashimoto A., Nagata Y., Fujii F., Kojima K., Suzuki K., Taya T., Tonouchi M., Nelson C., et al. Transcriptional profiling of genes responsive to abscisic acid and gibberellin in rice: Phenotyping and comparative analysis between rice and Arabidopsis. Physiol. Genomics. 2004;2:87–100. doi: 10.1152/physiolgenomics.00201.2003. [DOI] [PubMed] [Google Scholar]
- 63.Murphy A.M., Otto B., Brearley C.A., Carr J.P., Hanke D.E. A role for inositol hexakisphosphate in the maintenance of basal resistance to plant pathogens. Plant J. 2008;56:638–652. doi: 10.1111/j.1365-313X.2008.03629.x. [DOI] [PubMed] [Google Scholar]
- 64.Sreenivasulu N., Radchuk V., Strickert M., Miersch O., Weschke W., Wobus U. Gene expression patterns reveal tissue-specific signalling networks controlling programmed cell death and ABA-regulated maturation in developing barley grains. Plant J. 2006;47:310–327. doi: 10.1111/j.1365-313X.2006.02789.x. [DOI] [PubMed] [Google Scholar]
- 65.Thiel J., Weier D., Sreenivasulu N., Strickert M., Weichert N., Melzer M., Czauderna T., Wobus U., Weber H., Weschke W. Different hormonal regulation of cellular differentiation and function in nucellar projection and endosperm transfer cells: A microdissection-based transcriptome study of young barley grains. Plant Physiol. 2008;148:1436–1452. doi: 10.1104/pp.108.127001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmitteng L. Analysis of relative gene expression data using a real time quantitative PCR and 2-(Delta Delta C (T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 67.Gasparis S., Orczyk W., Zalewski W., Nadolska-Orczyk A. The RNA-mediated silencing of one of the Pin genes in allohexaploid wheat simultaneously decreases the expression of the other, and increases grain hardness. J. Exp. Bot. 2011;62:4025–4036. doi: 10.1093/jxb/err103. [DOI] [PubMed] [Google Scholar]
- 68.Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang X., Odom D.T., Koo S.H., Conkright M.D., Canettieri G., Best J., Chen H., Jenner R., Herbolsheimer E., Jacobsen E., Kadam S., Ecker J.R., Emerson B., Hogenesch J.B., Unterman T., Young R.A., Montminy M. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl. Acad. Sci. USA. 2005;22:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]