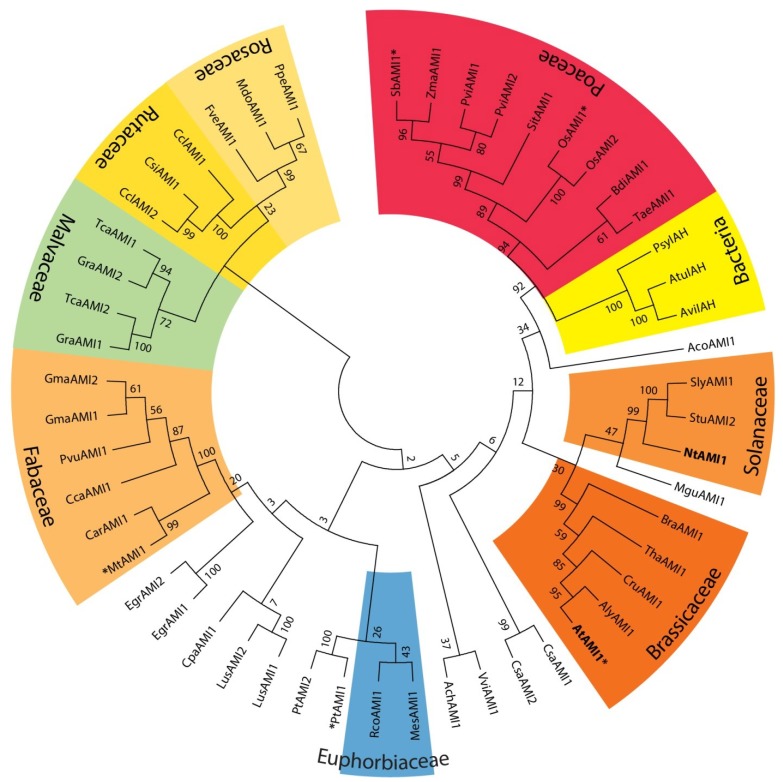
Figure 2.
Cladogram of 51 AMI1-like proteins from plants and bacteria.
A bootstrap consensus tree inferred from 500 replicates was generated to represent the phylogenetic relationships of the proteins analyzed. Proteins from the plant species highlighted in bold letters have already been enzymatically characterized in our group [26,30,31] or by other laboratories [27]. The amidases further studied in this work are labeled with asterisks. An amino acid sequence alignment of the selected enzymes can be taken from Appendix Figure A1. Plant species that belong to the same family are given in colored boxes. Species names are abbreviated as follows with gene/protein identifiers given in parenthesis: Ach: Actinidia chinensis (ACHN083681); Aco: Aquilegia coerulea (Aquca_05400135.1); Aly: Arabidopsis lyrata (16043473); At: Arabidopsis thaliana (At1g08980); Atu: Agrobacterium tumefaciens (YP_001967410); Avi: Agrobacterium vitis (P25016); Bdi: Brachypodium distachyon (Bradi5g27490); Bra: Brassica rapa (Bra031652); Car: Cicer arietinum (XP_004495736); Cca: Cajanus cajan (C.cajan06830); Ccl: Citrus clementine (AMI1: Ciclev10001239m, AMI2: Ciclev10001213m); Cpa: Carica papaya (evm.model.supercontig_48.34); Cru: Capsella rubella (Crubv10006860m); Csa: Cucumis sativus (AMI1: Cucsa.11590.1, AMI2: Cucsa.182140.1); Csi: Citrus sinensis (orange1.1g044294m); Egr: Eucalyptus grandis (AMI1: Eucgr.B01994.1, AMI2: Eucgr.B01996.1); Fve: Fragaria vesca (Mrna22572.1-v1.0-hybrid); Gma: Glycine max (AMI1: Glyma10g29640.1, AMI2: Glyma20g37660.1); Gra: Gossypium raimondii (AMI1: Gorai.007G295700.1, AMI2: Gorai.007G295900.1); Lus: Linum usitatissimum (AMI1: Lus10029941, AMI2: Lus10004466); Mdo: Malus x domestica (MDP0000302761); Mes: Manihot esculentum (cassava4.1_010118m); Mgu: Mimulus guttatus (mgv1a006860m); Mt: Medicago truncatula (Medtr1g082750.1); Nt: Nicotiana tabacum (AB457638); Os: Oryza sativa (AMI1: Os04g02780.1, AMI2: Os04g02754.1); Psy: Pseudomonas syringae (P52831); Ppe: Prunus persica (ppa005982m); Pt: Populus trichocarpa (AMI1: Potri.013g024100.1, AMI2: Potri.013g024200.1); Pvi: Panicum virgatum (AMI1: Pavirv00043873m, AMI2: Pavirv00038995m); Pvu: Phaseolus vulgaris (Phvul.007G180900.1); Rco: Ricinus communis (30128.m008759); Sb: Sorghum bicolor (Sb02g039510.1); Sit: Setaria italica (Si001473m); Sly: Solanum lycopersicum (Solyc10g086170.1.1); Stu: Solanum tuberosum (PGSC0003DMP400065555); Tae: Triticum aestivum (AK330138); Tca: Theobroma cacao (AMI1: Thecc1EG046817t1, AMI2: Thecc1EG046818t1); Tha: Thellungiella halophile (Thhalv10007679m); Vvi: Vitis vinifera (GSVIVT01000036001); Zma: Zea mays (GRMZM2G169087_T01).