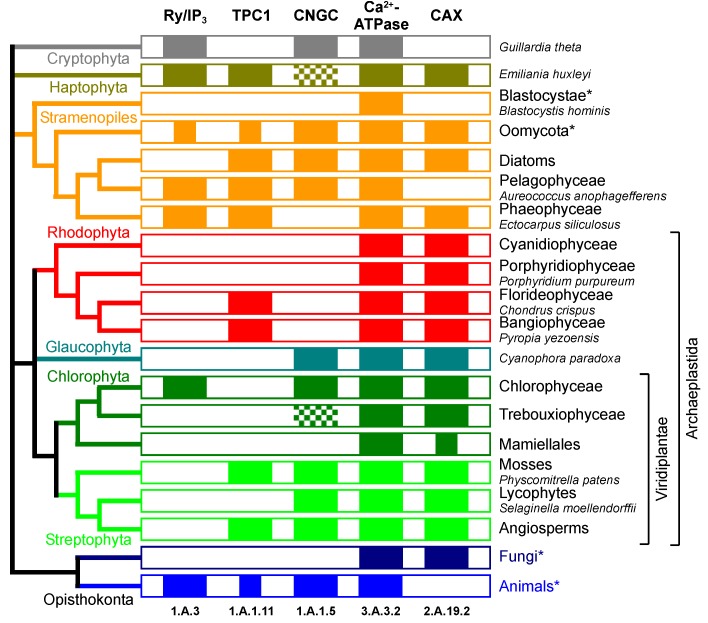
Figure 2.
Phylogenetic distribution of Ca2+ transporters in photosynthetic eukaryotes. Colored blocks indicate the presence of a certain Ca2+ transport protein (top and bottom labels) in a specific clade (labels on left and right). Abbreviations used for Ca2+ transport proteins: Ry/IP3, ryanodine-InsP3 receptor Ca2+ channel (TC 1.A.3); TPC1, two-pore Ca2+ channel (TC 1.A.1.11.13/18/19/22); CNGC, cyclic nucleotide-gated cation channel (TC 1.A.1.5); Ca2+-ATPase (TC 3.A.3.2); CAX, Ca2+:H+ exchanger (TC 2.A.19.2). Smaller block indicates that not all species in this clade seem to contain the Ca2+ transporter (see text). Checkered blocks indicate that it is currently not clear whether Haptophyta and Trebouxiphyceae contain cyclic nucleotide-gated cation channels or a similar cation channel from a different family of the voltage-gated ion channel superfamily (TC 1.A.1). The phylogenetic relationship (left [39]) of major eukaryotic photosynthetic clades (listed right) is presented and color coded (according to labels on the left). Clades marked with an asterisk are non-photosynthetic. For clades containing only one species with a fully sequenced genome, the species is given. Ca2+ transporters were identified by comparing established members from each Ca2+ transporter family to proteoms from selected eukaryotes with completely sequenced genomes using BLAST, followed by verification of each candidate by BLAST at TCDB [40,41].