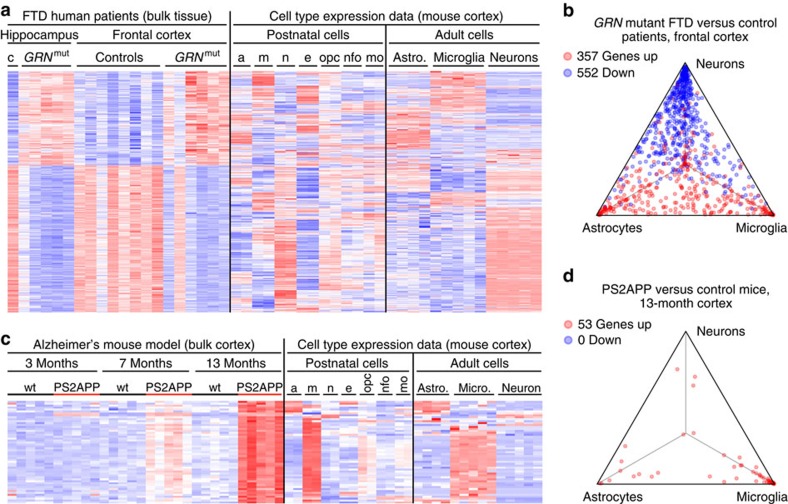
Figure 1. Mapping dementia-related gene expression profiles to CNS cell types.
(a) Genes differentially expressed by microarray analysis (fold change ⩾2, adjusted P≤0.05) in GRN mutant FTD patient cortex versus controls (GSE13162, imported) were analysed for expression in cerebrocortical cell types from normal postnatal (GSE52564, imported) or adult (GSE75246, this study) mouse brains. Each gene's expression was Z-score normalized separately within each of the three data sets (colour range: −4.10≤Z≤5.23). Rows (genes) were clustered hierarchically. Columns (RNA samples) were sorted by sample metadata. (b) The same genes with higher (red) or lower (blue) expression in GRN mutant FTD cortex are plotted in this triangle. A gene's proximity to a corner represents its degree of preferential expression in the indicated cell type from normal adult mouse cortex (see the Methods for details). (c,d) Similar to a,b, this heat map (colour range: −2.20≤Z≤3.47) and triangle plot analyse cell type expression for genes differentially expressed by microarray in bulk cerebral cortex from the PS2APP Alzheimer's model versus wild-type mice at 13 months of age (GSE74995, this study). a/astro., astrocytes; c, control; e, endothelial cells; m/micro., microglia; mo, myelinating oligodendrocytes; n, neurons; nfo, newly formed oligodendrocytes; opc, oligodendrocyte progenitor cells; wt, wild type.