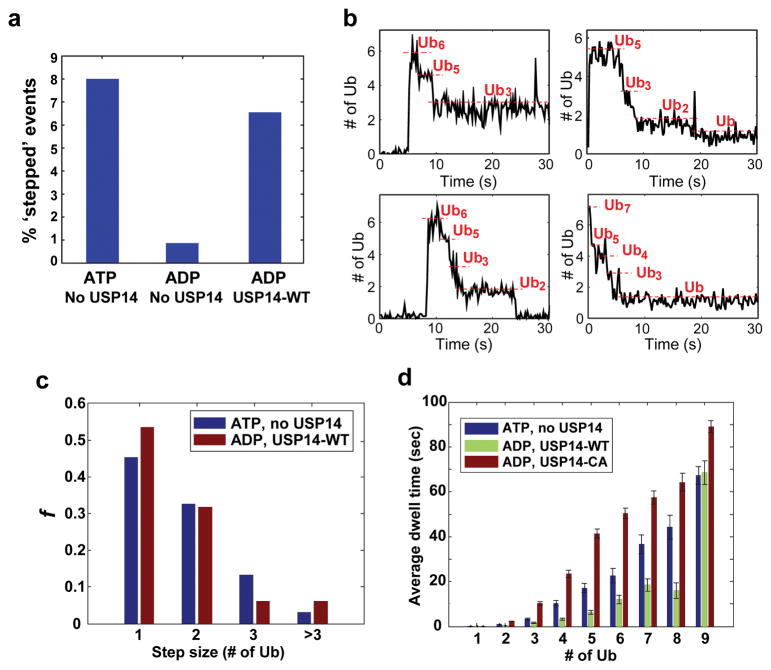
Figure 3. Single-molecule analysis of deubiquitination by USP14.
Single molecule (SM) assays were performed on immobilized human proteasome from which USP14 had been removed. Securin conjugates were generated using fluorescently labeled ubiquitin and added to the proteasome under conditions as indicated. a, Percent of stepped traces under conditions measuring RPN11 (ATP, no USP14) or USP14 (ADP, USP14-WT) activities. N(ATP, no USP14)=1574; N(ADP, no USP14)=578; N(ADP, USP14-WT)=733. b, Examples of step-trace segmentation graphs from an SM analysis on polyubiquitinated securin conjugates with USP14-reconstituted human proteasome in the presence of ADP (See Extended Data Fig. 10 for RPN11 step traces). c, Step size distribution of RPN11-mediated (ATP, no USP14, N=356) or USP14-mediated (ADP, USP14-WT, N=129) deubiquitination events as a function of the number of ubiquitins. d, Average dwell time of securin conjugates on the proteasome in the presence or absence of USP14. ATP was added for the no-USP14 condition. Dwell time was plotted as a function of the multiplicity of ubiquitin groups on securin. Error bars indicate SD of the mean (n=3). ADP denotes the condition of ADP plus o-PA to suppress RPN11 activity.