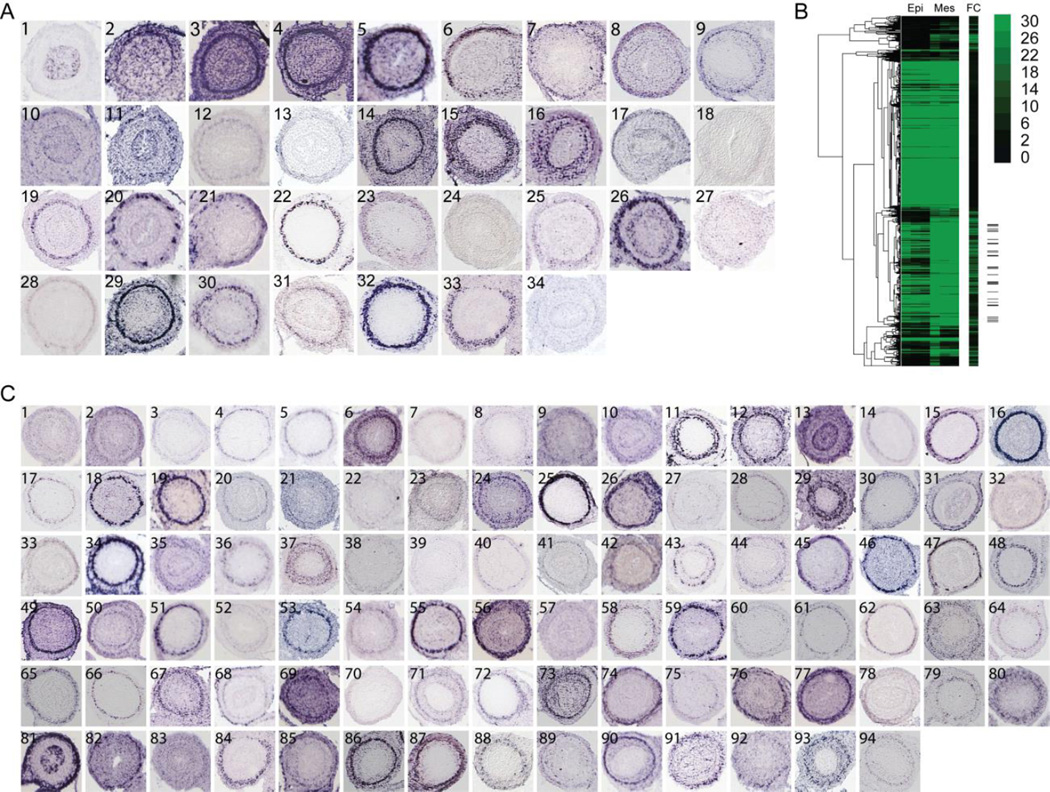
Figure 1. Identification of intestinal visceral smooth muscle genes.
(A) RNA in situ localization images of (A1) epithelial marker Cdx1 [EH4673] and (A2–34) GSEA identified prospective intestinal smooth muscle genes with ICM localization at E14.5. Intestinal cross sections are from Genepaint.org (Set ID in brackets). Panels are (2) ACTC1 [DC37], (3) ACTA2 [EH2333], (4) ACTG2 [ES688], (5) ADCY5 [MH851], (6) ATP1A2 [EN427], (7) BOC [EB1453], (8) CACNA1H [ES1420], (9) CALD1 [EG423], (10) CCL2 [EH401], (11) CNP1 [EH672], (12) DDR2 [EH3223], (13) DES [EH1766], (14) FLNA [EN2546], (15) FN1 [EN386], (16) FOXF2 [MH3070], (17) FXYD2 [EH804], (18) GLI3 [MH1026], (19) LMOD1 [EH4061], (20) MAPT [EH1455], (21) MEF2C [MH765], (22) MYL9 [EB2526], (23) NCAM1 [ES2731], (24) PLAU [MH1483], (25) PLCB4 [EH4161], (26) RBP1 [EH1743], (27) SALL2 [EH4737], (28) SLC8A1 [MH1892], (29) SMTN [EH1228], (30) SORBS1 [EH2043], (31) SVIL [EN1436], (32) TAGLN [EB415], (33) TGFBR2 [EB1113] and (34) TNNT2 [EH505] (B) Hierarchically clustered expression data containing three replicates of epithelial (Epi, first three columns) and mesenchyme (Mes, next three columns) read counts for genes upregulated in mesenchyme. Fold change (FC) is depicted in the additional column. The location of the confirmed smooth muscle genes (A2–34) are denoted on the right with black tick marks. (C) RNA in situ analysis of 94 other genes within the subcluster of confirmed GSEA enriched genes showing ICM localization. (1) ABI2 [EH4750], (2) ACOT7 [EH3555], (3) ADAM19 [EB509], (4) ADAMTS8 [EB1785], (5) ADCY5 [MH851], (6) AKAP2 [EH3708], (7) AKAP6 [EH3709], (8) AKAP12 [EB1789], (9) AKT3 [EH2186], (10) ATP2B4 [EG2318], (11) CAV1 [EG91], (12) CASQ1 [EN103], (13) CBX6 [EH1869], (14) CCKAR [EH3710], (15) CHRM2 [EH2853], (16) CNN1 [EH1511], (17) CRISPLD2 [ES1352], (18) CRMP1 [ES1162], (19) CTTNBP2 [EH1103], (20) EFS [EH2735], (21) CUEDC1 [EH1301], (22) DDR2 [EH3223], (23) DUSP10 [MH1605], (24) CSPR1 [HD13], (25) ENC1 [MH749], (26) FNBP1 [ES598], (27) FBXO32 [ES255], (28) FOXP2 [EG742], (29) EMILIN3 [EB1359], (30) FOXF1 [MH3518], (31) GDNF [EH574], (32) GAB2 [EN1340], (33) FREM2 [EN2329], (34) FZD3 [MH732], (35) GAS1 [MH360], (36) GEM [EH1664], (37) GNAQ [EN2590], (38) GPR20 [EH2873], (39) GLI1 [EN1215], (40) HAND1 [MH519], (41) GREM2 [EH1239], (42) GREM1 [EB63], (43) HOXB5 [EH612], (44) HHIP [EB1363], (45) HOXA5 [EN692], (46) HOXD3 [EN1290], (47) HOXB4 [MH3073], (48) ID4 [EN2437], (49) ITIH5 [EH3960], (50) ITGA9 [EH3560], (51) ITGA5 [EH3234], (52) KCNIP1 [EH1584], (53) LEBREL1 [EH4020], (54) IGFBP2 [ES388], (55) LRGI1 [MH431], (56) MYADM [EG746], (57) MTAP1B [ES1557], (58) MRVI1 [ES819], (59) LTBP1 [EH4243], (60) MAPK8IP1 [EH3404], (61) NEXN [ES414], (62) MYOCD [EN2445], (63) NCAM1 [ES2731], (64) MYOM1 [DC35], (65) NCALD [EG960], (66) NELL2 [EG1866], (67) NRBP2 [EH2160], (68) PSD [EB1667], (69) NPNT [MH2326], (70) PCDH7 [EB2060], (71) NKD1 [EB1514], (72) PKRG1 [EB1942], (73) PTN [EH3730], (74) PTMS [EG1747], (75) RGMA [EH2791], (76) RSPO3 [EN1484], (77) RARB [EH1089], (78) PYGL [MH3193], (79) SOX12 [MH3020], (80) SMOC2 [EH2269], (81) TGFB2 [EB1113], (82) SLC24A3 [MH3376], (83) SLC22A17 [MH2066], (84) SOX4 [MH3053], (85) TGFB1I1 [EG2803], (86) TGFB3 [EH2000], (87) TSPAN9 [EH2385], (88) THBS4 [MH1698], (89) THBS2 [ES844], (90) TMEM47 [EH2893], (91) TSHZ1 [EB2573], (92) TULP4 [MY240], (93) ZYX [EG1608], (94) WWTR1 [EH904)