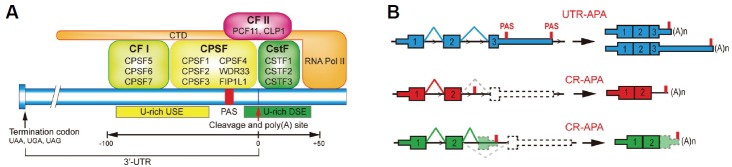
Fig. 1.
(A) 3′ end processing machinery assembly at poly(A) site. The endonucleolytic cleavage at poly(A) site is mediated by the binding of 3′ end processing factors including multi-subunit protein complexes CPSF, CSFT, CFI, and CFII to the USE-PAS-DSE sequence elements. (B) Schematic illustration of UTR-APA and CR-APA of mRNA. Exons are represented by rectangular squares with coding DNA sequences being thicker than UTRs. Introns are represented by thin lines. For UTR-APA, alternative PAS resides in 3′ UTR. Therefore, UTR-APA can generate transcripts with varying UTR lengths without changing the coding sequences. There are two major types of CR-APA, as illustrated. Both types of CR-APA may yield transcripts with truncated coding sequences.