Fig. 5.
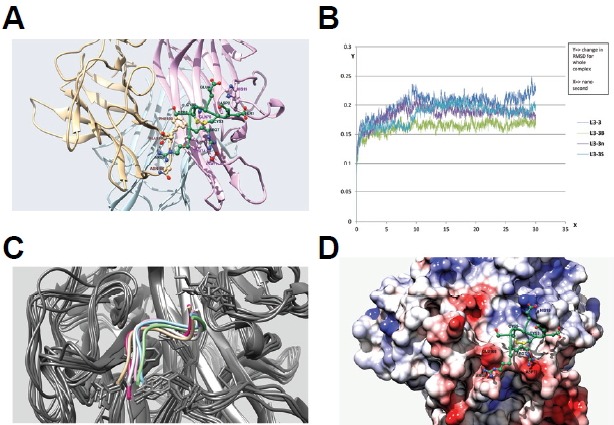
Docking and molecular dynamics simulation of RANKL inhibitory peptides. (A) Docking simulation of the L3-3B peptide. L3-3B is depicted in green, and monomers of the RANKL trimer are colored light brown, light pink, and sky blue. Residues are labeled using the same color scheme with darker shading. (B) Comparison of RMSD graphs from 30-ns molecular dynamics simulation results for four peptides: L3-3, L3-3B, L3-3n, and L3-3S. (C) Overlap of six coordinates of the L3-3B-RANKL complex captured over the course of a 50-ns molecular dynamics simulation. Different backbone orientations of the L3-3B peptide are shown in different colors, with all RANKL backbones colored grey. (D) Binding mode of the L3-3B peptide on the RANKL surface. The RANKL surface is mapped using Coulombic surface-coloring gradient (Chimera 1.10.1), where deep red and deep blue indicate surface charges of −10 and +10, respectively, and white indicates a neutral surface charge. The L3-3B peptide backbone is shown in green, and positive and negative side-chain atoms are depicted in blue and red, respectively. The C3–C5 disulfide bond is colored yellow.
