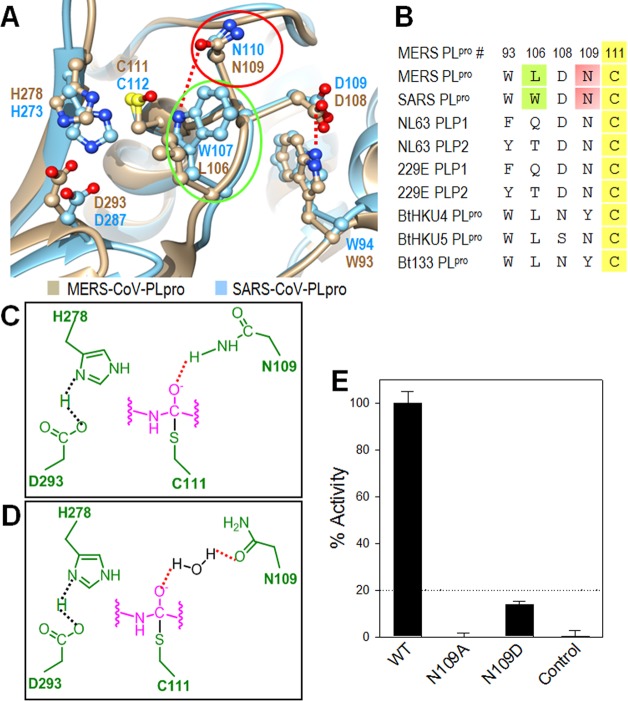
Figure 7.
Active site analysis of the MERS-PLpro. (A) Active site alignment of MERS-PLpro (tan) and SARS-PLpro (cyan). The three catalytic triad residues (C111, H278, and D293) of MERS-PLpro are aligned with the SARS-PLpro catalytic triad (C112, H273 and D287). (B) Sequence alignment of important residues near the catalytic triad between various CoV. Residue numbers are shown for MERS-PLpro. (C) Potential mechanism 1 for oxyanion hole stabilization via N109. Active site and substrate residues are shown in green and pink, respectively. (D) Potential mechanism 2 for oxyanion hole stabilization via N109. (E) Enzyme activity comparison between wild-type and two mutant MERS-PLpro enzymes.