Fig. 2.
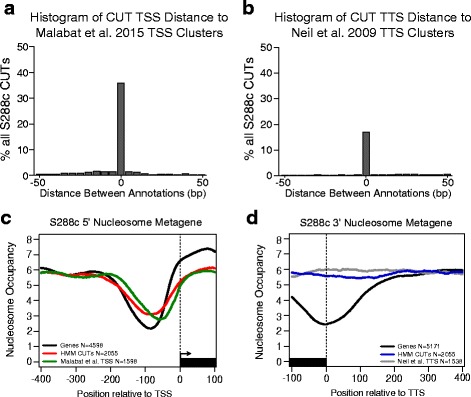
CUT start and stop sites concurrent with previous data and show distinct 3′ nucleosome structure. a Histogram showing the distribution of the distance between S288c CUT TSSs relative to Malabat et al. [32] CUT, intergenic, same sense, and antisense TSS clusters (see Methods). Histogram is only reporting distances for S288c CUTs that are within 50 bps of a TSS cluster. Bin widths are 5 bp. b Histogram showing the distribution of the distance between S288c CUT TSSs relative to Neil et al. [23] TTS clusters. Histogram is only reporting distances for S288c CUTs that are within 50 bps of a TTS cluster. Bin widths are 5 bp. c Metagene plot showing the average S288c nucleosome occupancy of a 500 bp window around the TSS for all genes with a 5′ UTR annotation (black), our HMM identified CUTs (red), and Malabat et al. [32] CUT TSS clusters (green). d Metagene plot showing the average S288c nucleosome occupancy of a 500 bp window around the TTS of all genes with a 3′ UTR annotation (black), our HMM identified CUTs (blue), and Neil et al. TTS clusters (grey)
