Extended Data Figure 7. Designs are stable to chemical denaturation by guanidine HCl (GuHCl).
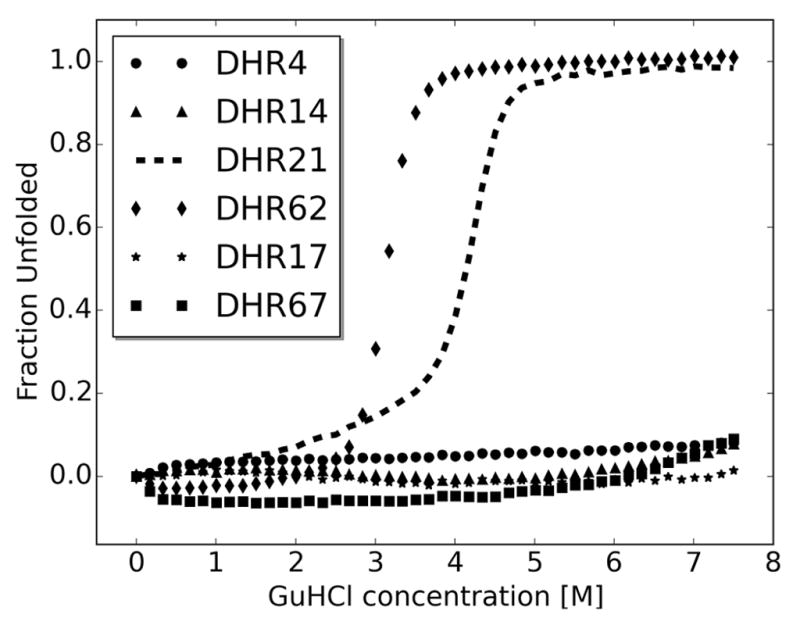
Circular dichroism monitored GuHCl denaturant experiments were carried for two designs for which crystal structures were solved (DHR4 and DHR14), two with overall shapes confirmed by SAXS (DHR21 and DHR62), and two with overall shapes inconsistent with SAXS (DHR17 and DHR67). In contrast to almost all native proteins, four of the six proteins do not denature at GuHCl concentrations up to 7.5 M. Both designs not confirmed by SAXS were extremely stable to GuHCl denaturation and hence are very well folded proteins; the discrepancies between the computed and experimental SAXS profiles may be due to small amounts of oligomeric species or variation in overall twist.
