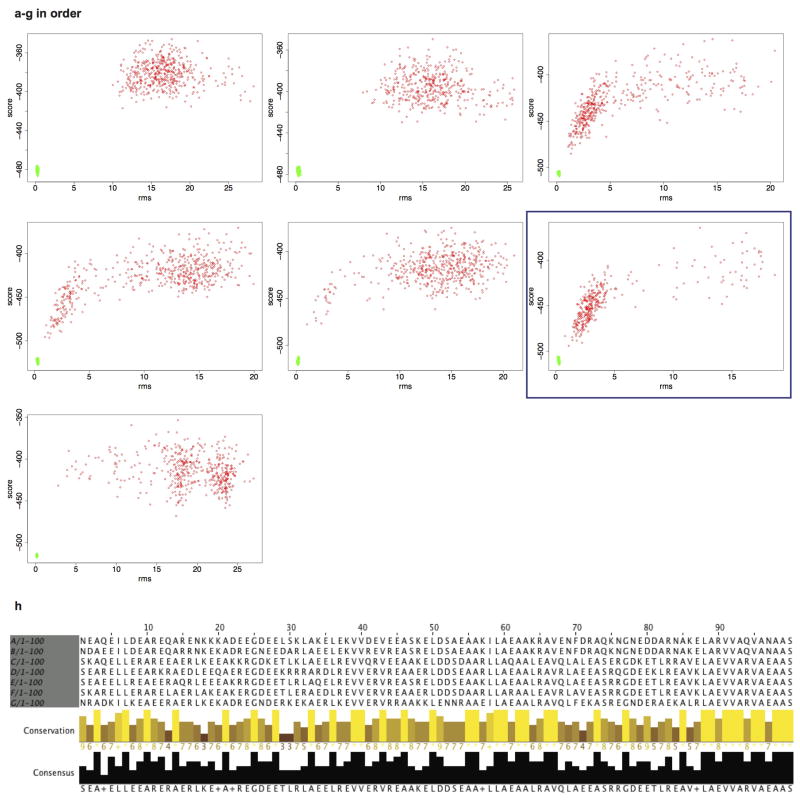
Extended Data Figure 3. Model validation by in silico folding.
To assess folding robustness seven sequence variants were made for each design. a-g illustrate the energy landscape explored by Rosetta ab-initio. In red are the protein models produced by ab initio search, in green by side chain repacking and minimization (relax). Models in deep global energy minima near the relaxed structures are considered folded. The variant with highest density of ab initio models near the relax region was chosen for experimental characterization (blue box). h, Jalview sequence alignment of the first 100 residues of the variants. The yellow bar height indicates sequence conservation, while the black bar how often the consensus sequence occurs.