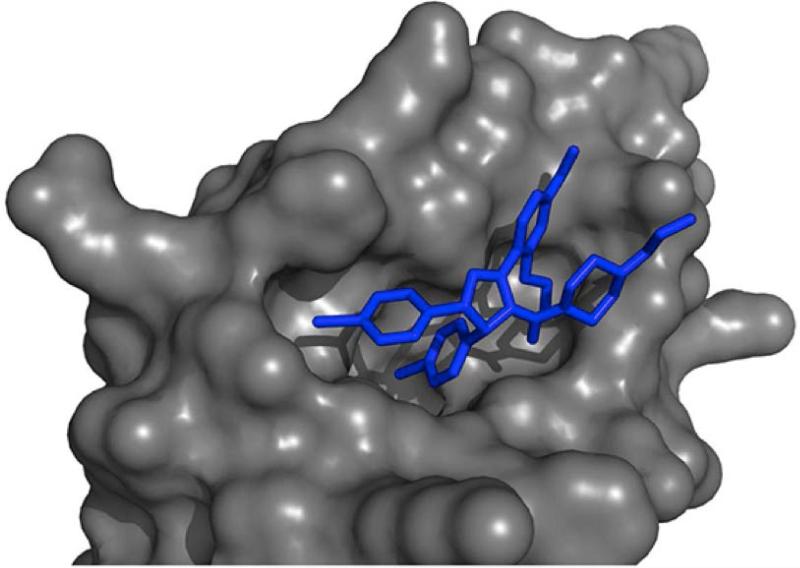
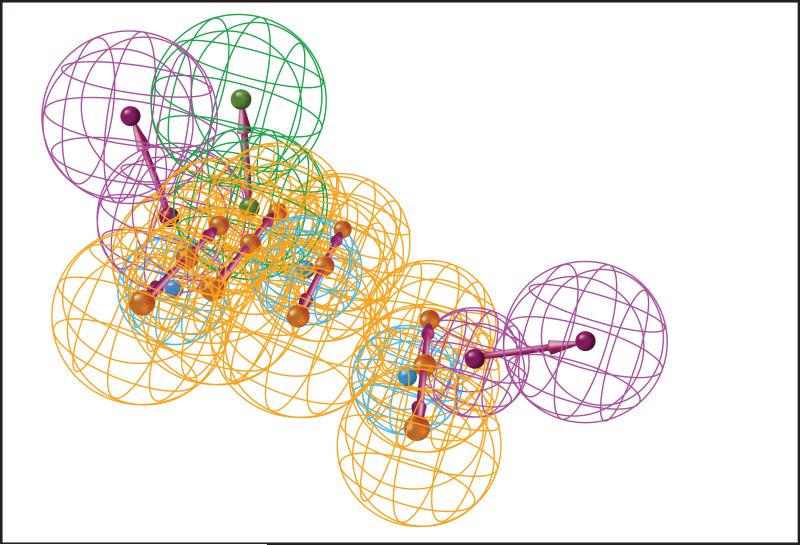
Figure 5. Computational chemistry approaches for target prediction.
5A: Result of a molecular docking simulation. The globular surface of the protein is shown in grey, and its docked ligand is in blue.
5B: Example of a pharmacophore model. A pharmacophore model is used to represent the chemical features deemed to be important for interaction with a chosen target. The features are arranged in three-dimensions along with some tolerance radius in an attempt to account for dynamic conformational changes of both protein and ligand. A pharmacophore model can be constructed from structural analysis of the target's binding pocket, or could be based on previously known interactions with the target. Compounds can then be aligned and scored against a pharmacophore model in order to prioritize likely interactions. Colors indicate different chemical descriptors such as hydrogen bond donor, or hydrogen bond acceptor, or hydrophobic region.