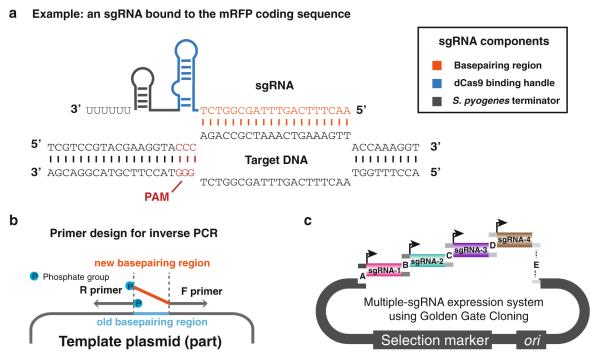
Fig. 1.
Design and cloning of sgRNAs for targeted gene repression. (a) Schematic of sgRNA binding to target DNA. The sgRNA consists of a 20-nt base-pairing region (orange), a 42-nt dCas9 binding handle (blue), and a 40-nt S. pyogenes transcription terminator (grey). The PAM is shown in red. Depicted is DNA from the coding sequence of RFP. (b) Schematic of generating vectors with new sgRNA sequences using inverse PCR. 5′ phosphorylation of oligos is indicated by blue circles. (c) Schematic of cloning multiple sgRNAs into a single vector using Golden Gate cloning