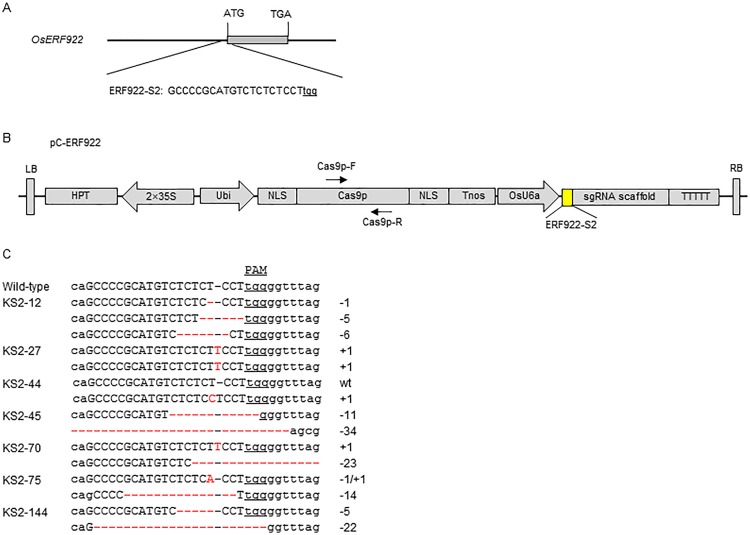
Fig 1. CRISPR/Cas9-induced OsERF922 gene modification in rice.
(A) Schematic of OsERF922 gene structure and the C-ERF922 target site (ERF922-S2). OsERF922 contains a single exon indicated by gray rectangles. The translation initiation codon (ATG) and termination codon (TGA) are shown. The target site nucleotides are shown in capital letters, and the protospacer adjacent motif (PAM) site is underlined. (B) Schematic diagram of the pC-ERF922 construct for expressing the CRISPR/Cas9 protein C-ERF922. The positions and orientations of the primers Cas9p-F and Cas9p-R, which were used to screen Cas9-free mutants, are indicated by small arrows. The expression of Cas9 is driven by the maize ubiquitin promoter (Ubi); the expression of the sgRNA scaffold is driven by the rice U6a small nuclear RNA promoter (OsU6a); the expression of hygromycin (HPT) is driven by 2 CaMV35S promoters (2 × 35S). NLS: nuclear localization signal; Tnos: gene terminator; LB and RB: left border and right border, respectively. (C) Nucleotide sequences at the target site in the 7 T0 mutant rice plants. The recovered mutated alleles are shown below the wild-type sequence. The target site nucleotides are indicated as black capital letters and black dashes. The PAM site is underlined. The red dashes indicate the deleted nucleotides. The red capital letters indicate the inserted nucleotides. The numbers on the right indicate the type of mutation and the number of nucleotides involved. “−” and “+” indicate the deletion and insertion of the indicated number of nucleotides, respectively; “−/+” indicates the simultaneous deletion and insertion of the indicated number of nucleotide.