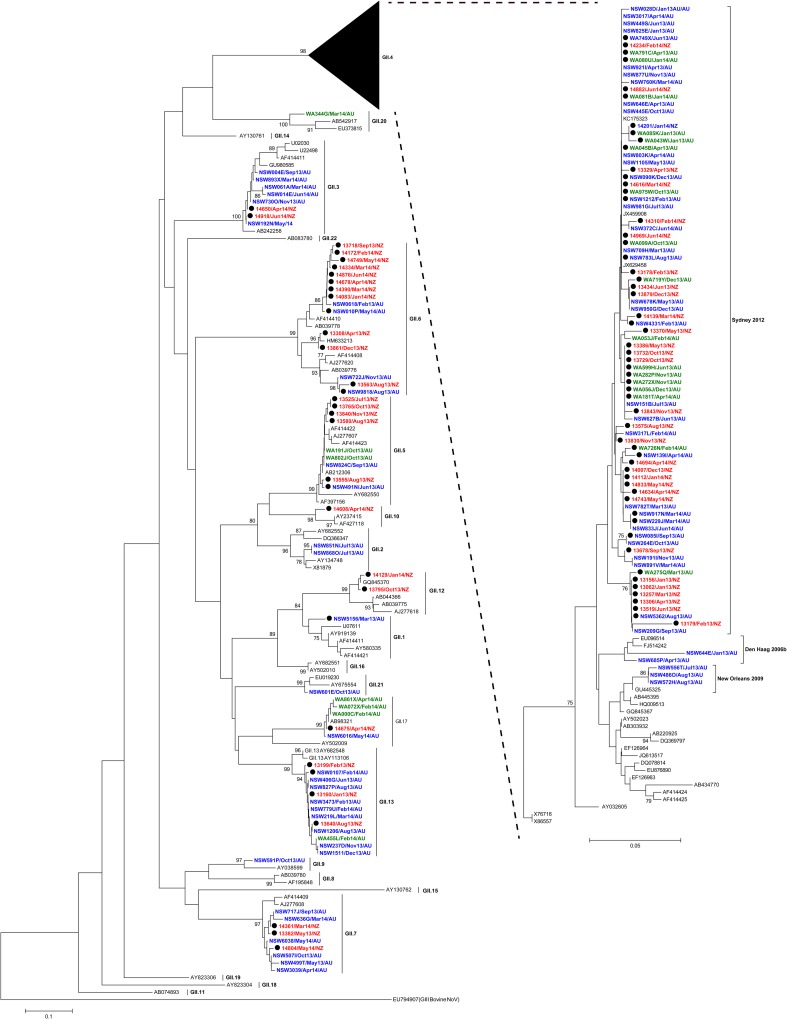
Fig 3. Phylogenetic analysis of NoV GII partial capsid nucleotide (nt) sequences from Australia and New Zealand.
Neighbour-Joining phylogeny of sequences of the 5’ end of ORF2 was generated. NoV GII sequences (266-bp, nt position 5101–5366 with reference to Lordsdale virus, GenBank accession number X86557) are shown. Global NoV reference sequences (n = 78) genotype were obtained from GenBank and are labelled with the GenBank accession number in black. The detailed reference strain information and collection year are tabulated in S1 Table. Representative NoV sequences used for the phylogenetic analysis (n = 156) are labelled with red (New Zealand), blue (NSW Australia) and green (WA Australia), in the format of: Sample ID/Collection month and year/Country. Each NoV-associated outbreak is indicated with a black solid circle. The phylogeny was generated using programs within MEGA 5, with bootstrap values of ≥75 indicated as a percentage of 1000 replicates. The distance scale represents the number of nucleotide substitutions per site. AU—Australia; NZ—New Zealand.