Figure 1.
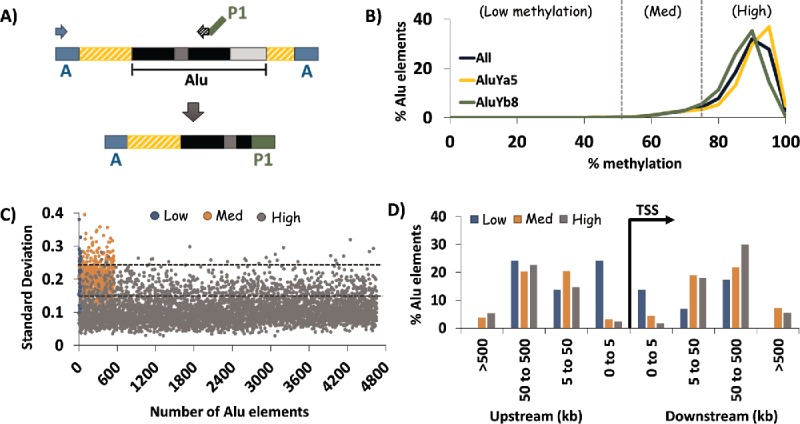
HT-TREBS of AluYa5 and AluYb8 in human fibroblast cells. (A) Primer scheme depicting the enrichment of AluYa5 and AluYb8 sequences using HT-TREBS. Briefly, methylated Ion Torrent “A” adaptors were ligated to sheared and size-selected genomic DNA, and the sample was bisulfite-treated. Ion “A” primers and subfamily-specific primers (with Ion “P1” adaptor on their 5′ ends) amplified fragments containing part of the Alu element (the first monomer and middle A-rich region) along with some flanking genomic sequence. (B) Methylation profile for AluYa5 and AluYb8 elements, binned in increments of 5%, for the 5238 loci sequenced. Loci were subdivided into 3 broad groups roughly corresponding to the inflection points in the graph: Low (0–50%), Medium (50–75%) and High methylation (75-100%). (C) Characterization of loci in terms of variation in methylation level (expressed in standard deviation, SD) for the 3 methylation groups. Dashed horizontal lines represent the thresholds used in the analysis (SD 0.15 and 0.25). Variation in DNA methylation was generally observed to be lower for highly methylated elements compared with those in the medium and low methylation groups. (D) Distance of AluYa5 andYb8 elements to the nearest transcription start site (TSS) based on methylation level. Alu elements with low levels of methylation appeared more likely to be present close to gene promoters compared with those methylated at >50% levels.
