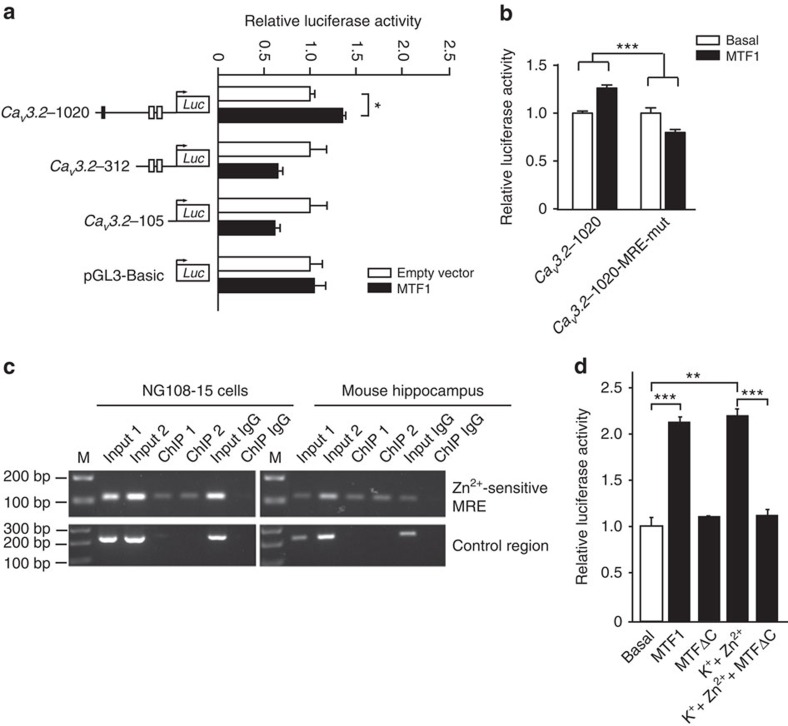
Figure 5. MTF1 binds the CaV3.2 promoter.
(a) Luciferase activity of the CaV3.2 promoter deletion fragments after overexpression with MTF1. Note the almost similar activation pattern of the CaV3.2 promoter deletion fragments as seen after stimulation with K++Zn2+ (50 mM/200 μM; Fig. 3c). No activation was observed for the pGL3-basic control plasmid (one-way analysis of variance (ANOVA): P=0.012; F(7,16)=3.856; Tukey's multiple comparisons test, *P≤0.05; n=3). (b) Luciferase activity of the CaV3.2-1020 fragment and the CaV3.2-1020 fragment with the mutated Zn2+-sensitive MRE-binding site (CaV3.2-1020-MRE-mut) after overexpression with MTF1. Mutation of the Zn2+-sensitive MRE-binding site resulted in a reduced CaV3.2 promoter activity (two-way ANOVA: P=0.0002; F(1,8)=38.9. (c) ChIP analysis of MTF1 binding to the Zn2+-sensitive MRE within the CaV3.2 promoter. PCR amplicons were generated of anti-MTF1 ChIP immunoprecipitates from NG108-15 cells and mouse hippocampi, using primer pairs spanning the Zn2+-sensitive MRE and a control region in the CaV3.2 promoter lacking a MRE. A rabbit-IgG immunoprecipitate was used as negative control. (d) Luciferase activity of unstimulated and K++Zn2+-challenged (50 mM/200 μM) NG108-15 cells transfected with the full-length CaV3.2 promoter–luciferase reporter construct and MTF1 or MTF1ΔC (one-way ANOVA: P<0.001; F(4,10)=117.9; Tukey's multiple comparisons test, **P≤0.01, ***P≤0.001; n≥3).