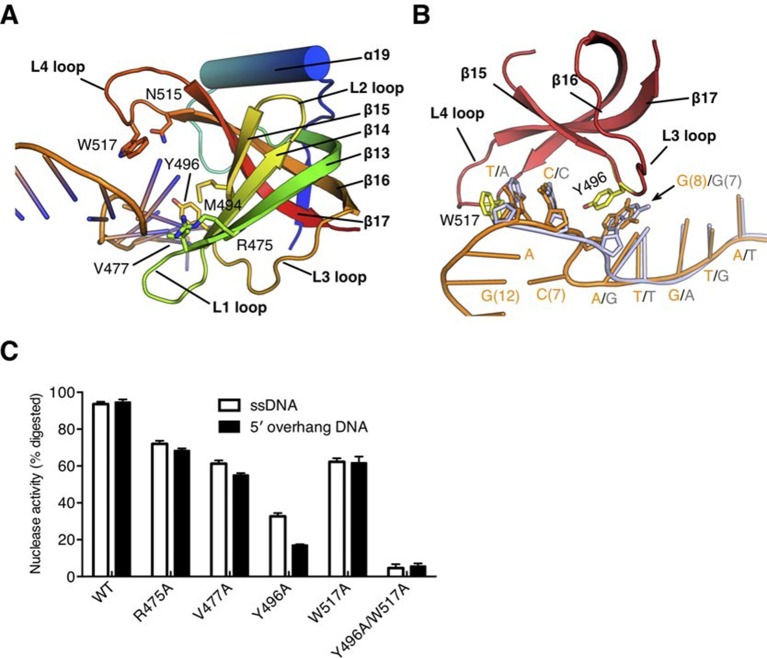
Figure 4. The OB fold domain is critical for drRecJ resection.
(A) The OB fold domain is shown in rainbow-colored diagrams. Residues involved in DNA binding are labeled and shown as sticks. (B) A comparison of DNA in the complex II (DNA with 5´-ssDNA overhang; orange) with complex III (ssDNA; white). Two conserved aromatic residues Tyr496 and Trp517 are shown as sticks (yellow). The black arrowhead indicates the position of stacking interaction between Tyr496 and the guanine base. (C) Quantification and plot of ssDNA and DNA with 5´-ssDNA overhang, which are processed by wild-type and mutant drRecJ proteins (alanine substitutions of key residues in the OB fold domain), using the same DNA substrate and reaction conditions as in Figure 1B. Data are represented as mean ± SEM.