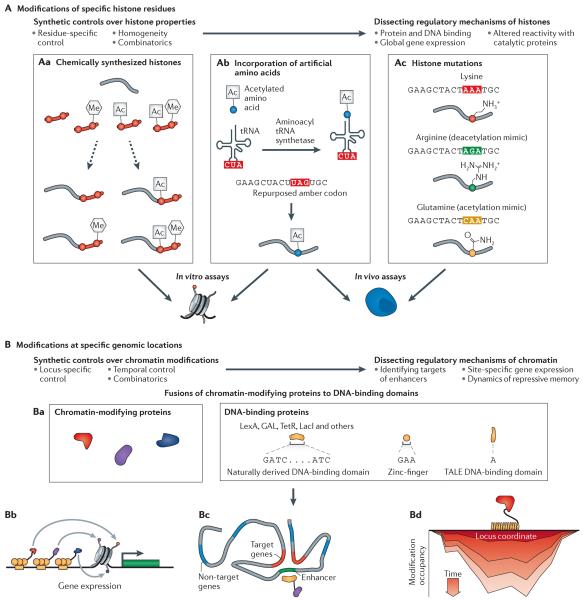
Figure 2. Synthetic control of biochemical chromatin modifications.
Synthetic approaches provide two types of specificity: specificity in the histone residues that are modified (part A) and specificity in the genomic locations of histone modifications (part B). Aa | Synthetic histones can be created by ligating chemically defined peptides (red) to the amino termini of partial histone proteins that are produced recombinantly (grey)30–32. Ab | Artificial amino acids that are already biochemically modified can be incorporated into specific residues of histones that are recombinantly produced in bacteria33–37. This approach requires expression of an aminoacyl tRNA synthetase and an acetyl-lysine transporter to import exogenously supplied acetyl-lysine. Ac | Genetic mutations can change specific residues into ones that partially mimic the charge and shape of a modified histone residue38–40. Histones that are chemically synthesized or that contain artificial amino acids can be used in in vitro assays to measure changes in binding affinities to regulatory proteins or altered reactivity of specific histone residues due to the presence of modifications on other residues. Histones that were genetically mutated can be used in cellular assays to measure the effects of specific residues on global gene expression profiles. Ba | Chromatin-modifying proteins with catalytic properties such as acetyltransferase or methyltransferase activities can be fused to DNA-binding proteins42–45,47,48,52–56. Bb–c | The fusion proteins bind to specific locations in the genome, altering gene expression of a downstream gene (part Bb) or genes that are targeted by a bound enhancer (part Bc). Bd | Chromatin modifiers can also be dynamically recruited to a genomic locus, allowing temporal measurements of changes in chromatin modifications53,66,70. TALE, transcription activator-like effector.