Fig. 6. crtS is crucial for Chr2 replication initiation at ori2.
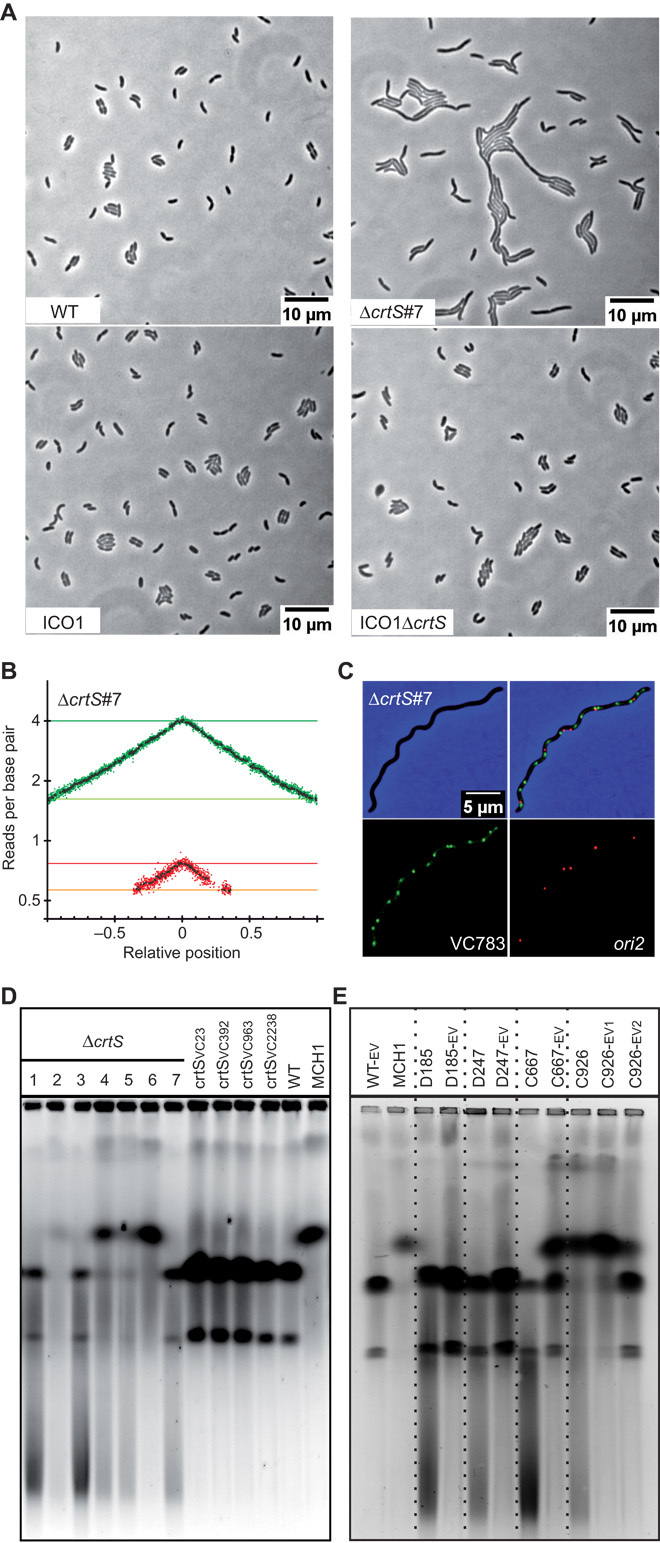
(A) Phenotype of crtS-deleted mutants in WT and ICO1. Representative pictures of phase-contrast microscopy of live cells growing on LB agar pads: WT, ΔcrtS #7 (with two separate chromosomes), ICO1, and ICO1ΔcrtS. (B) MFA of WTΔcrtS #7. (C) Representative picture of a filamentous ΔcrtS cell observed with fluorescence microscopy. Loci near crtS (VC783) and ori2 were fluorescently labeled. Merged pictures of VC783 (green) and ori2 (red) and phase-contrast (blue) micrographs show a higher number of green spots than red spots. (D) Ethidium bromide–stained pulsed-field gel electrophoresis (PFGE) of native gDNA. From left to right: Independent clones of crtS-deleted mutants (WTΔcrtS #1 to #7), mutants with relocated crtS (crtSVC23, crtSVC392, crtSVC963, and crtSVC2238), and WT (two chromosomes) and MCH1 (one synthetic fused chromosome), which are used as size reference (18). (E) Same as (D). From left to right: WT-EV (control), MCH1 (one synthetic fused chromosome), and independent clones of ΔcrtS mutants before (D185, D247, C667, and C926) and after a 200-generation evolution (-EV). For C926, samples were harvested after 100 (-EV1) and 200 generations (-EV2) to track the tendency of the fused chromosome to revert to two separate chromosomes.
