Summary
Centromeres control genetic inheritance by directing chromosome segregation but are not genetically encoded themselves. Rather, centromeres are defined by nucleosomes containing CENP-A, a histone H3 variant [1]. In cycling somatic cells, centromere identity is maintained by an established cell cycle-coupled CENP-A chromatin assembly pathway, but how centromeres are inherited through the mammalian female germline is unclear because of the long (months to decades) prophase I arrest. We show that mouse oocytes retain the pool of CENP-A nucleosomes assembled before birth, and this pool is sufficient for centromere function, fertility, and genome transmission to embryos. Indeed, oocytes lack any measurable CENP-A nucleosome assembly through the entire fertile lifespan of the female (>1 year). Thus, the remarkable stability of CENP-A nucleosomes confers transgenerational centromere identity in mammals.
Results & Discussion
A pathway for centromere inheritance between somatic cell cycles is well established [1]. CENP-A nucleosomes redistribute equally between sister centromeres during DNA replication, and newly synthesized CENP-A is assembled at human centromeres exclusively in early G1-phase of the cell cycle [2, 3]. Therefore, centromere inheritance depends on retention of CENP-A nucleosomes from incorporation into chromatin in S-phase until new loading in the subsequent G1-phase. This cell cycle-coupled mechanism of centromere maintenance raises the question of how centromeres are inherited between generations in the mammalian female germline. Between pre-meiotic S-phase in the oocyte and the subsequent G1-phase in the zygote, mammalian oocytes arrest in an extended prophase I that may last from a few months to decades depending on species [4]. Three attractive models could explain how centromere identity is maintained during this meiotic arrest: 1) CENP-A is not retained at centromeres during female meiosis, and centromere identity in the germline is independent of CENP-A, 2) CENP-A is maintained through the action of a meiotic chromatin assembly pathway, which is distinct from the timing of the established cell cycle-coupled pathway in cycling somatic cells, or 3) CENP-A nucleosomes assembled at oocyte centromeres are extremely stable and maintain centromere identity in the absence of any nascent CENP-A chromatin assembly during the prolonged arrest. The first two models have strong precedent in holocentric worms [5, 6] and flies [7], respectively. The third model is supported by the notable stability of CENP-A nucleosomes in chromatin: they do not measurably redistribute between centromeres during an entire cell cycle, and their turnover in rapidly dividing somatic mammalian cells is so slow that it can be explained by dilution through segregation on daughter strands of replicating centromere DNA [2, 3, 8]. We use mouse as a model system to distinguish between these models for centromere inheritance in the mammalian female germline.
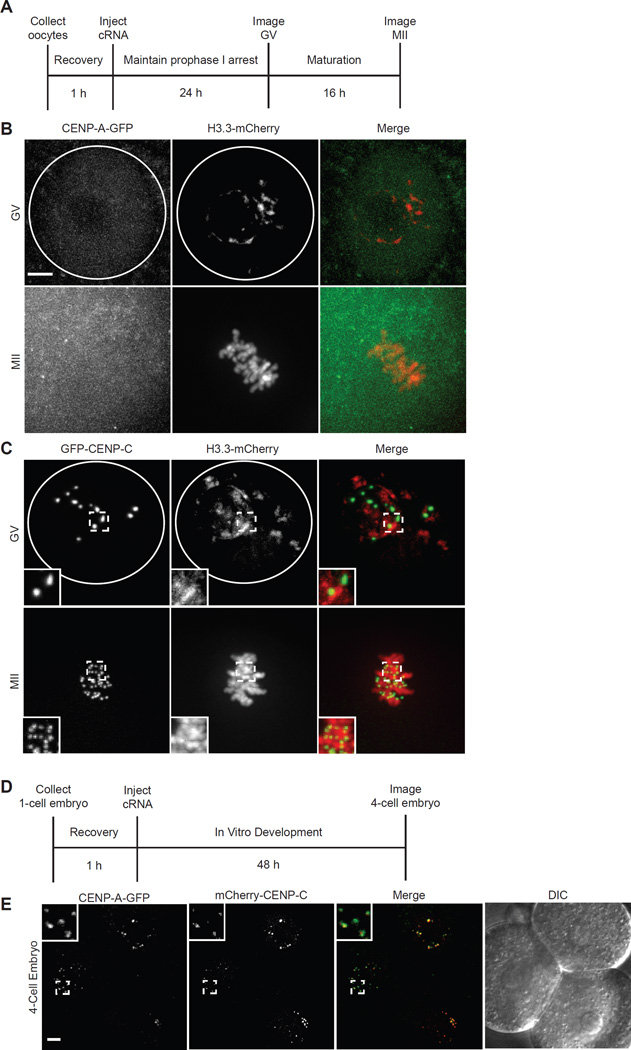
We first tested for a meiotic CENP-A chromatin assembly pathway on a short timescale in full grown germinal vesicle intact (GV) oocytes by injecting cRNAs encoding fluorescently tagged proteins: CENP-A, the histone H3 variant H3.3, or CENP-C, which binds CENP-A nucleosomes at centromeres [2, 9] (Fig. 1A). We find that H3.3-mCherry assembles into chromatin, as previously shown in oocytes [10, 11], and GFP-CENP-C targets to centromeres, but no CENP-A-GFP is detected at centromeres either in prophase of meiosis I (GV) or after maturation to metaphase II (MII; Fig. 1B,C), though we do detect a measureable increase in GFP fluorescence in oocytes injected with CENP-A-GFP cRNA (Fig. S1A). The CENP-A-GFP cRNA produced functional protein capable of assembly at centromeres, as shown by co-localization of CENP-A-GFP and mCherry-CENP-C in 4-cell embryos (Fig. 1D,E). We obtained similar results using CENP-A tagged with Flag and HA epitopes instead of GFP (Fig. S1B,C). These data show that a meiosis-specific loading pathway does not assemble measurable CENP-A in full grown oocytes on a timescale of ~40 h.
Figure 1. Absence of CENP-A chromatin assembly in full grown oocytes or MII eggs.
(A–C) Oocytes were injected with cRNA for CENP-A-GFP and H3.3-mCherry or GFP-CENP-C and H3.3-mCherry, and imaged live in germinal vesicle-intact (GV) or metaphase II (MII) stages as shown in the schematic (A). Representative images show CENP-A-GFP and H3.3-mCherry (B) or GFP-CENP-C and H3.3-mCherry (C) at GV or MII stages (n≥10 in each case). Images are maximal intensity projections of confocal z-series; insets show 1.6x magnification of centromeres labeled with GFP-CENP-C. White circle represents nuclear envelope. (B) 1-cell stage embryos were injected with cRNA for mCherry-CENP-C and CENP-A-GFP and imaged live at the 4-cell stage, as shown in the schematic (D). Representative images show an optical section of 3 blastomeres (n=27); insets show 3x magnification of CENP-A-GFP and mCherry-CENP-C localized at centromeres. Scale bars 5 µm. See also Figure S1.
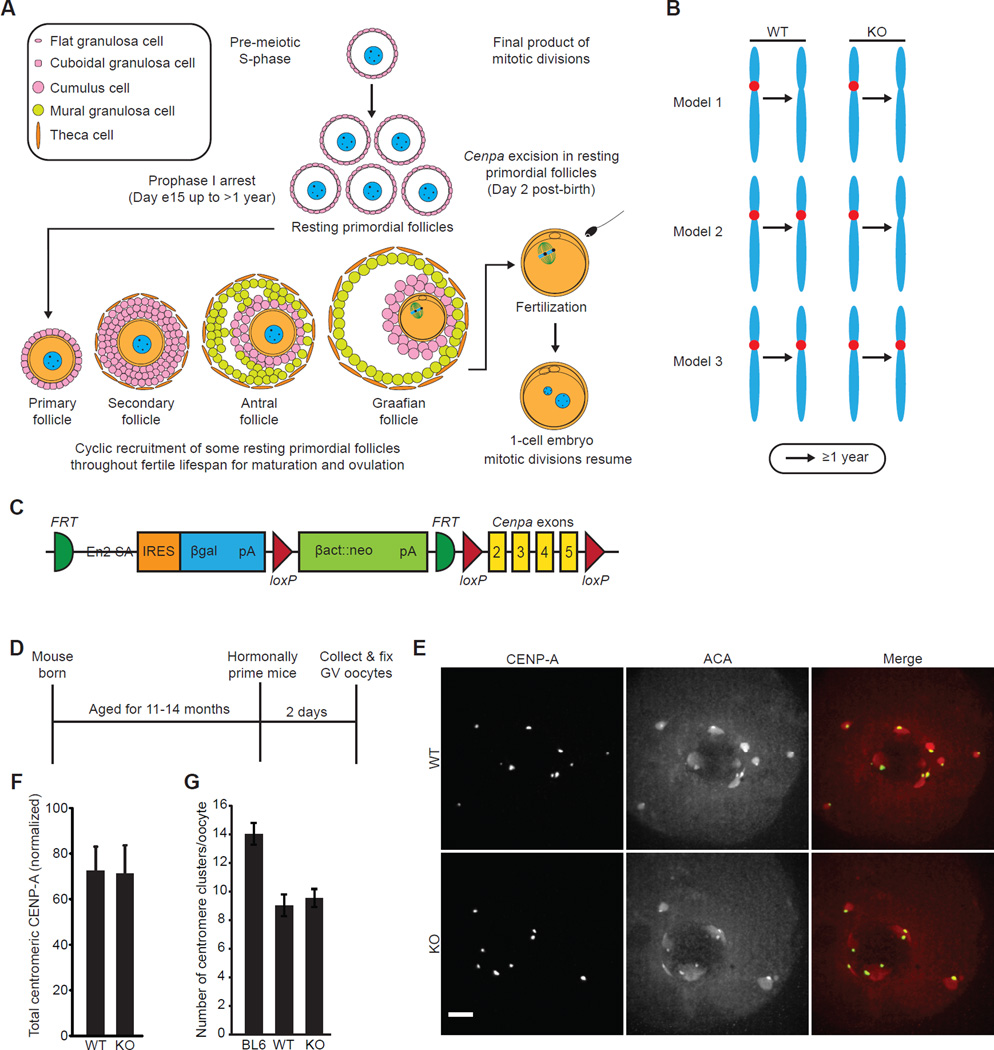
To test for CENP-A nucleosome assembly on timescales of many months during the prophase I arrest, we designed a genetic experiment in which we knocked out Cenpa in oocytes in resting primordial follicles (Fig. 2A). Oocytes in these animals would rely on CENP-A nucleosomes that were assembled at centromeres prior to the knockout. Therefore, if CENP-A is simply lost at oocyte centromeres, as in the worm (Fig. 2B, model 1) [5, 6], then CENP-A should become undetectable in both knockout and control oocytes. Alternatively, if centromere inheritance depends on meiotic assembly of nascent CENP-A nucleosomes, then centromeric CENP-A levels should decrease in the knockout oocytes over time (Fig. 2B, model 2). Finally, if centromere inheritance relies on the stable retention of preassembled CENP-A nucleosomes, then we expect little change in centromeric CENP-A protein levels over time (Fig. 2B, model 3). Because a pool of long-lived CENP-A protein in the oocyte would confound interpretation of our gene deletion experiment, we measured total CENP-A protein in oocytes as compared to cycling NIH 3T3 cells. We found that 500 oocytes (4n) have approximately the same amount of CENP-A protein as ~1000 NIH 3T3 cells (with each cell between 3–6n, because they are near-tetraploid and from an asynchronous cycling population) (Fig. S2A). These data exclude the possibility that a large pool of excess CENP-A protein exists in oocytes to replenish CENP-A at centromeres in the absence of new synthesis.
Figure 2. CENP-A nucleosomes are stably retained at oocyte centromeres for >1 year.
(A) Oocytes arrest in prophase I in resting primordial follicles, which are cyclically recruited starting five days after birth to begin growth towards a Graafian follicle, the final stage before ovulation. Primordial follicles can remain in the resting phase for a period lasting >1 year before they are recruited for maturation and ovulation. After fertilization, the maternal and paternal pronuclei enter G1-phase and begin mitotic cell cycles. Cre expression driven by the Gdf9 promoter excises Cenpa in resting primordial follicles two days after birth. (B) Three models for centromere inheritance in the mammalian oocyte. See Results & Discussion for details. (C) Schematic of Cenpa conditional knockout gene locus. The neomycin selection cassette used for selection of ES cells is flanked by FRT sites. Cenpa protein coding exons 2–5 are flanked by loxP sites. (D–F) Oocytes were collected from 11–14.5 month old WT and KO mice, or from young C57BL/6J (BL6) controls, and CENP-A levels were analyzed by immunofluorescence, with ACA to co-label centromeres (schematic, D). Images (E) of oocytes with intact germinal vesicles (GV) are maximal intensity projections of confocal z-series; scale bar 5 µm. Total centromeric CENP-A staining was quantified for each oocyte (n=64 oocytes from 4 WT mice; n=85 oocytes from 5 KO mice) and normalized to young C57BL/6J controls (n=155 oocytes, 8 mice) for each experiment. Normalized values were averaged over multiple experiments (F; error bars, s.d.). (G) The number of centromere clusters was counted in each oocyte and averaged over each group (error bars, s.d.). See also Figures S2, S3, S4, and Table S1.
To conditionally knock out Cenpa, we generated a mouse carrying a Cenpa allele in which exons 2–5 are flanked by loxP sites (Fig. 2C) and used cre-recombinase driven by the Gdf9 promoter to inactivate Cenpa in oocytes in resting primordial follicles [12]. Multiple lines of evidence indicate that the Gdf9 promoter is active in these oocytes at an early stage. First, our microarray data show a high level of Gdf9 mRNA in isolated oocytes from resting primordial follicles collected two days after birth (day 2) [13]. Second, in an experimental system in which β-gal expression depends on Gdf9-Cre expression, primordial follicles stain positive at day 12 [12]. Because the first wave of follicle development begins at day 5, the β-gal positive oocytes in these primordial follicles must be in the resting state. Third, Gdf9-Cre mediated deletion of the Tsc1 or Pten genes, which are essential to maintain quiescence of primordial follicles, leads to activation of the entire pool of primordial follicles by day 23 [14, 15]. Finally, Gdf9-Cre mediated expression of Diptheria toxin leads to complete loss of oocytes by 8 weeks of age [16]. We derived Cenpafl/fl;Gdf9-Cre− (WT) or Cenpafl/fl;Gdf9-Cre+ (KO) females in a C57BL/6J background (Fig. S2B, Table S1). To confirm that the Cenpa gene locus is deleted in KO oocytes, we measured Cenpa mRNA levels by analyzing cDNA libraries created by reverse transcription of pooled mRNA from KO or WT oocytes (Fig. S3A). We found that KO oocytes from young animals (14–19 weeks) contain only trace amounts of Cenpa mRNA (0.7% ± 0.3%) relative to that present in WT oocytes (Fig. S3B,C). Additionally, we detected measurable differences in Cenpa mRNA when cDNA from KO and WT oocytes was mixed in the following ratios: 100% KO, 90% KO/10% WT, and 95% KO/5% WT (Fig. S3D,E). These data demonstrate that the Cenpa locus is efficiently excised in all KO oocytes and exclude the possibility of a stored pool of long-lived Cenpa mRNA.
Now positioned to test models for how centromere identity is maintained in mammalian oocytes, we measured centromeric CENP-A levels in the oocytes of WT and KO mice after aging them for 11–14.5 months (Fig. 2D–F). Centromeres are highly clustered in young oocytes and become even more clustered with age (Fig. 2G). We measured total centromeric CENP-A fluorescence in each oocyte (Fig. S3F–H) and normalized the signal to that in oocytes obtained from young C57BL/6J controls, to compare biologically replicated experiments (Fig. 2F). We find that aged KO and WT oocytes are indistinguishable, inconsistent with nascent CENP-A nucleosome assembly during the prolonged prophase I arrest (Fig. 2B, model 2). Similarly, cohesion is established in pre-meiotic S-phase without detectable cohesin turnover during prophase I [17]. Further, both the WT and KO groups retain ~70% of the CENP-A protein seen in the young C57BL/6J group, showing that the majority of CENP-A present since early prophase I arrest in the primordial follicle is retained at the centromere for ~1 year. This retention is also inconsistent with a model where CENP-A is removed from centromeres in oocytes and then centromere identity is reestablished in the early embryo, as in worms (Fig. 2B, model 1) [5, 6]. These findings are, however, entirely consistent with a model (Fig. 2B, model 3) where CENP-A nucleosomes assembled at the centromere before the prophase I arrest maintain centromere identity and function for the fertile lifespan of the female.
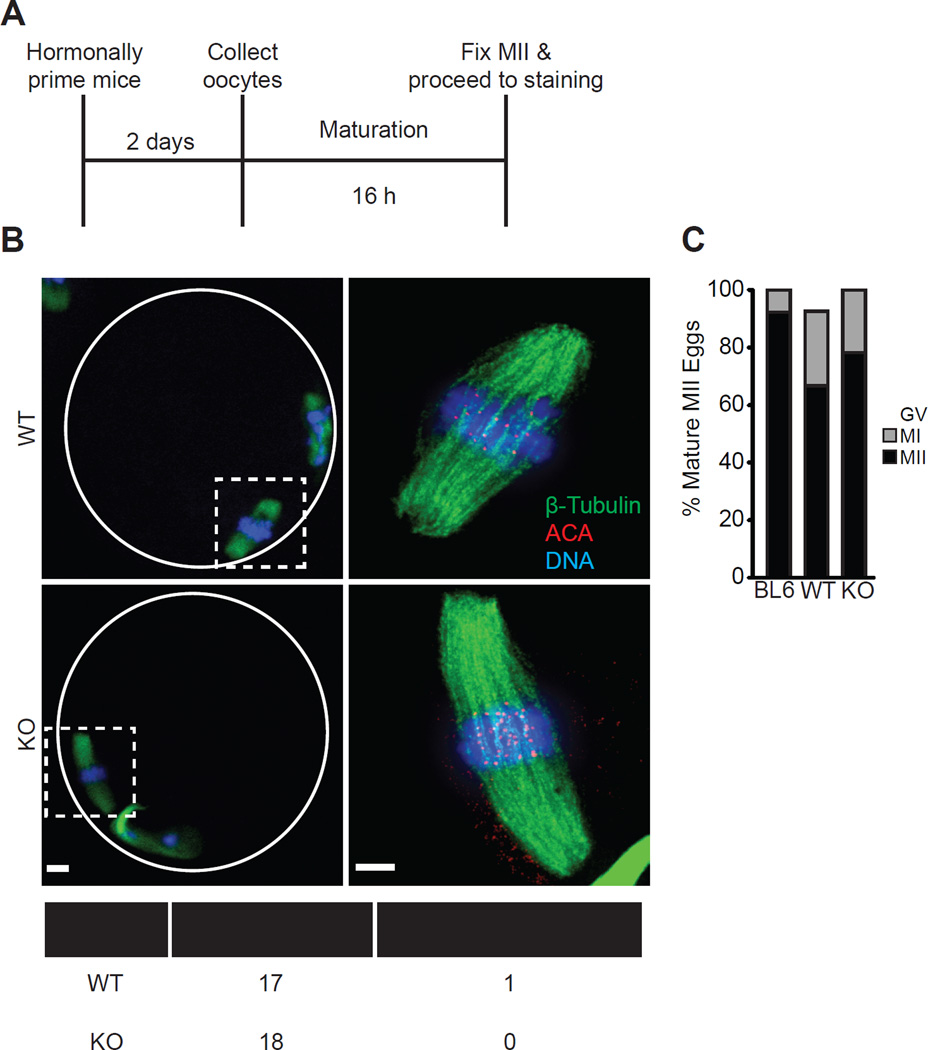
A broadly conserved feature of eukaryotic centromeres is the essential role of CENP-A nucleosomes in building a functional kinetochore [18–23]. To test whether the persistence of CENP-A nucleosomes at the centromere, rather than persistent expression, suffices for its essential function in the oocyte, we measured progression to metaphase II (MII) and chromosome alignment on MII spindles (Fig. 3A). Oocytes collected from 12 month old KO and WT mice were similar in both assays, consistent with our finding that centromeric CENP-A levels are equivalent between the two groups (Fig. 3B,C). Fewer oocytes from the 12 month groups matured to MII eggs compared to young C57BL/6J controls, consistent with age-related meiotic defects [24–26].
Figure 3. CENP-A nucleosomes assembled early in prophase I support normal meiotic centromere function.
(A–C) Oocytes were collected from 12 month old WT and KO females, or young C57BL/6J controls, matured in vitro to MII, and stained for β-tubulin, ACA to label centromeres, and DNA (schematic, A). Representative images (B) show MII eggs at 20X magnification (scale bar 10 µm), white circle represents zona pellucida; dashed squares are regions imaged at 100x (scale bar 5 µm). The percent in each group that remained arrested with an intact germinal vesicle (GV), arrested at metaphase I, or progressed normally to MII was quantified (C, n=27 oocytes from 3 WT mice, 23 oocytes from 3 KO mice, or 26 oocytes from 2 C57BL/6J (BL6) mice). The table summarizes how many MII eggs from WT and KO females had chromosomes aligned at metaphase.
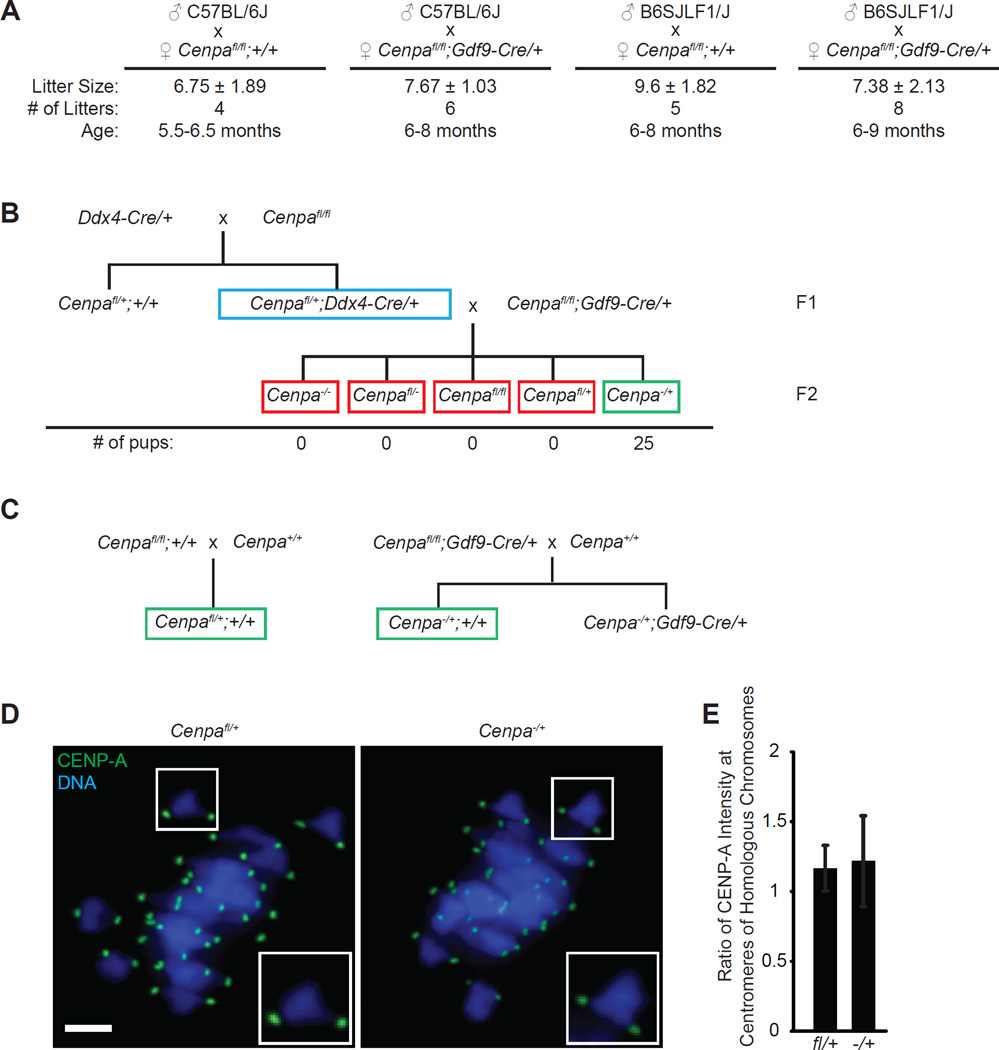
As the ultimate test of centromere function after Cenpa deletion in oocytes, we measured fertility of WT and KO females. We used 5–9 month old females, as older animals (WT or KO) have prohibitively low fecundity and thus yield too few data points. We find that fertility is indistinguishable between WT and KO females, indicating that CENP-A nucleosomes assembled at centromeres in oocytes before the prophase I arrest provide full fertility months later (Fig. 4A). Prior to receiving CENP-A protein, message, and/or gene from the WT sperm, meiotic divisions in the oocyte would be completed with the CENP-A nucleosomes assembled 5–9 months earlier.
Figure 4. CENP-A nucleosomes assembled in early prophase I support normal fertility and transgenerational centromere inheritance.
(A) WT (n=5) and KO (n=6) females were mated with either C57BL/6J or B6SJLF1/J males to determine fertility. Average litter size (± s.d.), number of litters, and age of the females are reported. (B) Cenpafl/+;Ddx4-Cre/+ males (blue box) were mated to Cenpafl/fl;Gdf9-Cre/+ females (n=2 females, 3–6 months old). The red box represents offspring that are either predicted to die in utero (all Cenpa−/−) or are predicted to be impossible to produce if there is full excision of the Cenpafl allele. The green box represents animals that are predicted to survive (all Cenpa−/+) if all oocytes are indeed Cenpa−/−. Surviving pups were genotyped and found to be exclusively Cenpa−/+ with no intact Cenpafl allele. (C) Breeding scheme to generate F1 pups that are Cenpafl/+ or Cenpa−/+. Green boxes indicate mice used in experiment. (D,E) Oocytes from F1 pups were fixed in metaphase I and stained for CENP-A and DNA. CENP-A intensity was measured at each centromere, and the ratio of brighter/dimmer intensity was calculated (E) for each bivalent (n≥238 bivalents for WT and KO). Error bars s.d. Scale bar 5 µm.
Our findings demonstrate that Cenpa deletion early in the prophase I arrest has no effect on oocyte maturation or female fertility. To independently verify that the Cenpa locus was deleted, we designed a cross in which survival of half of the offspring would depend on the presence of an intact maternal Cenpa allele in the egg (Fig. 4B). We used the Ddx4 promoter, which turns on in primordial germ cells, to excise a floxed Cenpa allele in spermatogonial stem cells [27]. Thus, Cenpafl/+;Ddx4-Cre/+ males produce mature sperm that are either Cenpa− or Cenpa+. If Cenpa is deleted in oocytes from KO females, all eggs produced will be Cenpa−. Crossing a KO female with a Cenpafl/+;Ddx4-Cre/+ male should produce embryos that are either Cenpa−/+ or Cenpa−/−, depending on the sperm genotype. Cenpa−/+ animals are viable and fertile, but Cenpa−/− is an embryonic lethal phenotype [20], so the surviving offspring from the cross should all be Cenpa−/+ and the litter sizes should be small. If Cenpa is not deleted in all oocytes, some offspring will be Cenpafl/+ or Cenpafl/−. We find that pups generated from this cross are exclusively Cenpa−/+ (Fig. 4B), confirming that the Cenpa locus is excised in all KO female oocytes. The viability of the Cenpa−/+ pups provides additional evidence that the CENP-A nucleosomes present at centromeres in KO oocytes are sufficient to produce a viable MII egg and support early embryogenesis.
To determine whether CENP-A nucleosomes present early in the prophase I arrest are sufficient for transgenerational inheritance of centromere identity, we examined the offspring of KO females crossed to normal C57BL/6J males. Defects in centromere inheritance between the maternally and paternally inherited chromosomes would be most apparent in metaphase I of the offspring, when the homologous chromosomes are paired. We crossed WT or KO females with C57BL/6J males, collected oocytes from F1 pups that were either Cenpafl/+ or Cenpa−/+ (Fig. 4C), and matured them to metaphase I in vitro. On each bivalent, we measured CENP-A levels on each side (i.e., at the centromeres of the homologous chromosomes) by immunofluorescence. If centromere inheritance were partially impaired, we would expect asymmetric CENP-A staining on each bivalent, which would imply that the maternal centromeres inherited less CENP-A. We find that it is symmetric, however, and the ratio of CENP-A staining within each bivalent is close to one for both genotypes (Fig. 4D,E). Thus, faithful transgenerational centromere inheritance is maintained by the stable retention of CENP-A nucleosomes through the extended prophase I arrest of the oocyte.
Conclusions
Taken together, our results provide clear genetic evidence that stable retention of CENP-A nucleosomes underlies centromere inheritance through the mammalian female germline. CENP-A protein incorporated into centromeric chromatin before the prophase I arrest, most likely in the G1-phase preceding meiotic entry, displays spectacular longevity and is sufficient to provide essential centromere function for >1 year. This mechanism of centromere inheritance represents a new paradigm when compared to what has been described in other organisms [5–7]. Our results stand in stark contrast to recent reports in which deposition of another histone H3 variant, H3.3, during prophase I is required for normal chromatin structure and gene expression and oocyte survival [11, 28] Thus, centromeric chromatin remains extremely stable while nucleosome assembly in bulk chromatin is ongoing and essential during the extended prophase I arrest. Pericentromeric heterochromatin (e.g. immunostaining for HP1β and H3K9me3; Fig. S4) is similar between oocytes and NIH 3T3 cells, so we prefer the notion that the stability we observe in the oocyte is a property conferred by CENP-A nucleosomes [29], themselves, and/or other components of the centromere [2, 30]. In addition to the important implications for understanding how centromere identity is transmitted transgenerationally in mammals, our findings also advance what we know about the functions of long-lived proteins. Many of the most remarkable examples are from metabolically inactive environments (e.g., crystallin and collagen [31, 32]) or of some nuclear components of non-dividing neurons [33–35]. Our findings extend the role of long-lived proteins to one that plays a central and essential role in the orchestration of chromosome segregation in the quintessential pluripotent cell type. At the centromere, the remarkable stability of CENP-A nucleosomes through the fertile lifespan of the female cements the epigenetic information required to faithfully guide chromosome inheritance and transmit the chromosomal location of the centromere to offspring.
Supplementary Material
Acknowledgments
We thank J. Bell for technical assistance and J. Thorvaldsen and the UPenn Transgenic and Chimeric Mouse Facility for assistance in generating the Cenpa conditional knockout mice. This work was supported by a predoctoral fellowship from AHA (E.M.S). We acknowledge support from NIH grants GM008275 (UPenn Structural Biology Training Grant; E.M.S.), HD058730 (R.M.S., M.A.L., and B.E.B.), a pilot grant from the UPenn Center for Study of Epigenetics in Reproduction HD068157 (M.A.L. and B.E.B.), a pilot grant from the UPenn Epigenetics Program (B.E.B. and M.A.L.), and a pilot grant from the UPenn Institute on Aging (M.A.L., B.E.B., and R.M.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
E.M.S., P.S., R.M.S., M.A.L., and B.E.B. designed the experiments. E.M.S. and P.S. conducted the experiments. E.M.S., P.S., R.M.S., M.A.L., and B.E.B. analyzed the data. E.M.S., M.A.L. and B.E.B. wrote the manuscript.
Contributor Information
Michael A. Lampson, Email: lampson@sas.upenn.edu.
Ben E. Black, Email: blackbe@mail.med.upenn.edu.
References
- 1.Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-A nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falk SJ, Guo LY, Sekulic N, Smoak EM, Mani T, Logsdon GA, Gupta K, Jansen LET, Van Duyne GD, Vinogradov SA, et al. CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science. 2015;348:699–704. doi: 10.1126/science.1259308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jansen LET, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J. Cell. Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Von Stetina JR, Orr-Weaver TL. Developmental control of oocyte maturation and egg activation in metazoan models. Cold Spring Harb. Perspect. Biol. 2011;3:1–19. doi: 10.1101/cshperspect.a005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monen J, Maddox PS, Hyndman F, Oegema K, Desai A. Differential role of CENP-A in the segregation of holocentric C. elegans chromosomes during meiosis and mitosis. Nat. Cell Biol. 2005;7:1248–1255. doi: 10.1038/ncb1331. [DOI] [PubMed] [Google Scholar]
- 6.Gassmann R, Rechtsteiner A, Yuen KW, Muroyama A, Egelhofer T, Gaydos L, Barron F, Maddox P, Essex A, Monen J, et al. An inverse relationship to germline transcription defines centromeric chromatin in C. elegans. Nature. 2012;484:534–537. doi: 10.1038/nature10973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunleavy EM, Beier NL, Gorgescu W, Tang J, Costes SV, Karpen GH. The cell cycle timing of centromeric chromatin assembly in Drosophila meiosis is distinct from mitosis yet requires CAL1 and CENP-C. PLoS Biol. 2012;10:1–16. doi: 10.1371/journal.pbio.1001460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodor DL, Valente LP, Mata JF, Black BE, Jansen LET. Assembly in G1 phase and long-term stability are unique intrinsic features of CENP-A nucleosomes. Mol Biol Cell. 2013;24:923–932. doi: 10.1091/mbc.E13-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato H, Jiang J, Zhou B-R, Rozendaal M, Feng H, Ghirlando R, Xiao TS, Straight AF, Bai Y. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science. 2013;340:1110–1113. doi: 10.1126/science.1235532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akiyama T, Suzuki O, Matsuda J, Aoki F. Dynamic replacement of histone H3 variants reprograms epigenetic marks in early mouse embryos. PLoS Genet. 2011;7:1–12. doi: 10.1371/journal.pgen.1002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nashun B, Hill PWS, Smallwood SA, Dharmalingam G, Amouroux R, Clark SJ, Sharma V, Ndjetehe E, Pelczar P, Festenstein RJ, et al. Continuous histone replacement by Hira is essential for normal transcriptional regulation and de novo DNA methylation during mouse oogenesis. Mol. Cell. 2015;60:611–625. doi: 10.1016/j.molcel.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lan Z-J, Xu X, Cooney AJ. Differential oocyte-specific expression of Cre recombinase activity in GDF-9-iCre, Zp3cre, and Msx2Cre transgenic mice. Biol. Reprod. 2004;71:1469–1474. doi: 10.1095/biolreprod.104.031757. [DOI] [PubMed] [Google Scholar]
- 13.Pan H, O’Brien MJ, Wigglesworth K, Eppig JJ, Schultz RM. Transcript profiling during mouse oocyte development and the effect of gonadotropin priming and development in vitro. Dev. Biol. 2005;286:493–506. doi: 10.1016/j.ydbio.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 14.Adhikari D, Zheng W, Shen Y, Gorre N, Hämäläinen T, Cooney AJ, Huhtaniemi I, Lan Z-J, Liu K. Tsc/mTORC1 signaling in oocytes governs the quiescence and activation of primordial follicles. Hum. Mol. Genet. 2010;19:397–410. doi: 10.1093/hmg/ddp483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reddy P, Liu L, Adhikari D, Jagarlamudi K, Rajareddy S, Shen Y, Du C, Tang W, Hämäläinen T, Peng SL, et al. Oocyte-specific deletion of Pten causes premature activation of the primordial follicle pool. Science. 2008;319:611–613. doi: 10.1126/science.1152257. [DOI] [PubMed] [Google Scholar]
- 16.Uhlenhaut NH, Jakob S, Anlag K, Eisenberger T, Sekido R, Kress J, Treier A-C, Klugmann C, Klasen C, Holter NI, et al. Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell. 2009;139:1130–1142. doi: 10.1016/j.cell.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 17.Burkhardt S, Borsos M, Szydlowska A, Godwin J, Williams SA, Cohen PE, Hirota T, Saitou M, Tachibana-Konwalski K. Chromosome Cohesion Established by Rec8-Cohesin in Fetal Oocytes Is Maintained without Detectable Turnover in Oocytes Arrested for Months in Mice. Curr. Biol. 2016 doi: 10.1016/j.cub.2015.12.073. Online Publication Date: February 18, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Black BE, Jansen LET, Maddox PS, Foltz DR, Desai AB, Shah JV, Cleveland DW. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol. Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 19.Goshima G, Kiyomitsu T, Yoda K, Yanagida M. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 2003;160:25–39. doi: 10.1083/jcb.200210005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howman EV, Fowler KJ, Newson AJ, Redward S, MacDonald AC, Kalitsis P, Choo KHA. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl. Acad. Sci. U.S.A. 2000;97:1148–1153. doi: 10.1073/pnas.97.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Régnier V, Vagnarelli P, Fukagawa T, Zerjal T, Burns E, Trouche D, Earnshaw W, Brown W. CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol. Cell Biol. 2005;25:3967–3981. doi: 10.1128/MCB.25.10.3967-3981.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- 23.Ravi M, Chan SWL. Haploid plants produced by centromere-mediated genome elimination. Nature. 2010;464:615–618. doi: 10.1038/nature08842. [DOI] [PubMed] [Google Scholar]
- 24.Subramanian VV, Bickel SE. Aging predisposes oocytes to meiotic nondisjunction when the cohesin subunit SMC1 is reduced. PLoS Genet. 2008;4:1–12. doi: 10.1371/journal.pgen.1000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lister LM, Kouznetsova A, Hyslop LA, Kalleas D, Pace SL, Barel JC, Nathan A, Floros V, Adelfalk C, Watanabe Y, et al. Age-related meiotic segregation errors in mammalian oocytes are preceded by depletion of cohesin and Sgo2. Curr. Biol. 2010;20:1511–1521. doi: 10.1016/j.cub.2010.08.023. [DOI] [PubMed] [Google Scholar]
- 26.Chiang T, Duncan FE, Schindler K, Schultz RM, Lampson MA. Evidence that weakened centromere cohesion is a leading cause of age-related aneuploidy in oocytes. Curr. Biol. 2010;20:1522–1528. doi: 10.1016/j.cub.2010.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gallardo T, Shirley L, John GB, Castrillon DH. Generation of a germ cell-specific mouse transgenic cre line, Vasa-Cre. Genesis. 2007;417:413–417. doi: 10.1002/dvg.20310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang MCW, Jacobs SA, Mattiske DM, Soh YM, Graham AN, Tran A, Lim SL, Hudson DF, Kalitsis P, O'Bryan MK, et al. Contribution of the two genes encoding histone variant H3.3 to viability and fertility in mice. PLoS Genet. 2015;11:1–23. doi: 10.1371/journal.pgen.1004964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 30.Falk SJ, Lee J, Sekulic N, Sennett MA, Lee T-H, Black BE. CENP-C directs a structural transition of CENP-A nucleosomes mainly through sliding of DNA gyres. Nat. Struct. Mol. Biol. 2016 doi: 10.1038/nsmb.3175. Online Publication Date: February 15, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masters PM, Bada JL, Zigler JS., Jr Aspartic acid racemisation in the human lens during ageing and in cataract formation. Nature. 1977;268:71–73. doi: 10.1038/268071a0. [DOI] [PubMed] [Google Scholar]
- 32.Verzijl N, DeGroot J, Thorpe SR, Bank RA, Shaw JN, Lyons TJ, Bijlsma JWJ, Lafeber FPJG, Baynes JW, TeKoppele JM. Effect of collagen turnover on the accumulation of advanced glycation end products. J. Biol. Chem. 2000;275:39027–39031. doi: 10.1074/jbc.M006700200. [DOI] [PubMed] [Google Scholar]
- 33.Commerford SL, Carsten AL, Cronkite EP. Histone turnover within nonproliferating cells. Proc. Natl. Acad. Sci. U.S.A. 1982;79:1163–1165. doi: 10.1073/pnas.79.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toyama BH, Savas JN, Park SK, Harris MS, Ingolia NT, Yates JR, III, Hetzer MW. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell. 2013;154:971–982. doi: 10.1016/j.cell.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Savas JN, Toyama BH, Xu T, Yates JR, Hetzer MW. Extremely long-lived nuclear pore proteins in the rat brain. Science. 2012;335:942. doi: 10.1126/science.1217421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.