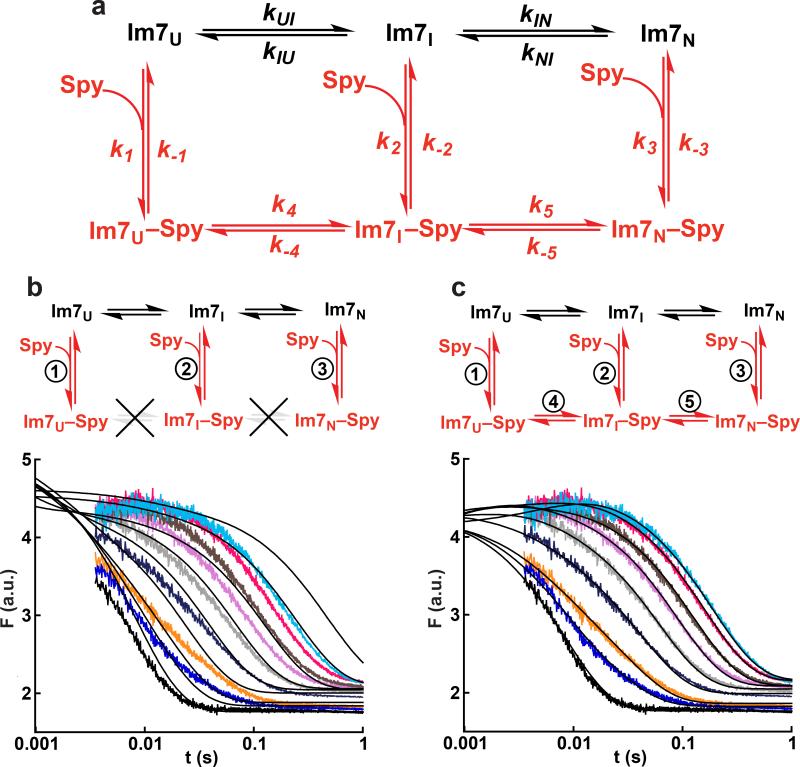
Figure 5.
Global fitting of Im7+Spy kinetic data. (a) The global fitting was built upon the well-characterized mechanism15–20 for Im7 folding in the absence of Spy (black path). The experimental fluorescence traces for Spy binding to Im7-WT and Im7-WT folding in the presence of Spy (Fig. 4) were globally fit to different mechanisms containing various combinations of the steps in red. In the global fitting, the equilibrium constant for the Im7U to Im7I step (KUI = kUI/kIU) and the forward and reverse rate constants for the Im7I to Im7N step were fixed to the values determined from the urea dependence of Im7 folding in the absence of Spy (Supplementary Fig. 5). (b, c) Attempted global fitting to the mechanism that omits and allows, respectively, the folding of Im7 while bound to Spy. For clarity, only the traces for Im7 folding in the presence of Spy are shown. The black lines in the plots are the best fit to the data. The mechanism that completely omits folding of Im7 while bound to Spy (b) fails to fit the data, whereas the mechanism that allows folding of Im7 while bound (c) can successfully fit the data. Global fitting to additional mechanisms and the best fit for the Spy binding Im7-WT data can be found in Supplementary Fig. 6.