Figure 1.
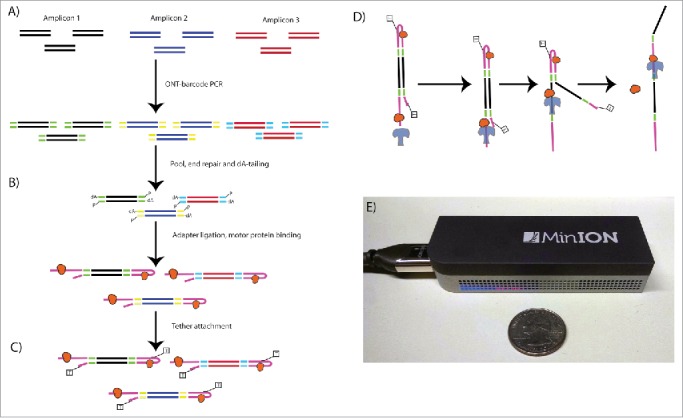
Nanopore Library Prep Workflow. Oxford Nanopore barcodes were incorporated into amplicons by PCR- individually for each SV- then resultant reactions were pooled (A). After NEB End Repair and dA-tailing modules (B), hairpin and leader adapters were ligated on, each containing a motor protein. Only the hairpin protein contained a his-tag, which was used to enrich for molecules containing a leader adapter and his-tag (his-tag selection step not shown). Tether attachment (C) allowed for direct attachment of the molecules to the flow cell membrane. Within the MinION flowcell (D), DNA molecules are pulled through a protein pore (blue), with motor protein (orange) affecting speed of DNA translocation through the pore. One side of the DNA molecule is read, then the hairpin, then the second side. Both reads were aligned to produce a 2D consensus read.
