Figure 1.
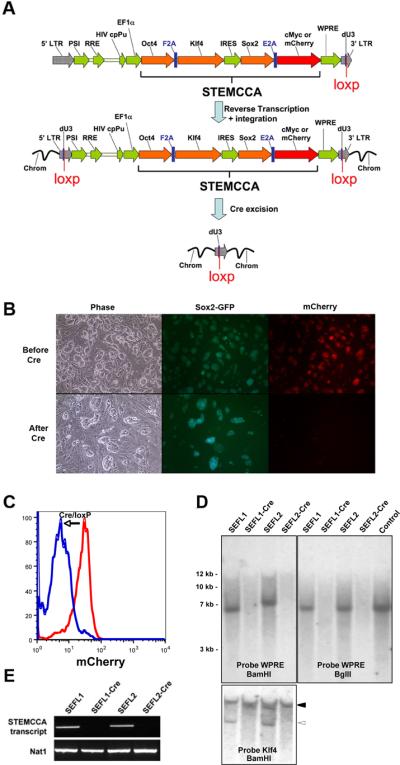
An excisable single lentiviral vector for the generation of iPS cells free of exogenous transgenes. (A) Schematic representation of the STEMCCA-loxP or STEMCCA-loxP-RedLight lentiviral vector. Two versions of the constitutive EF1α-STEMCCA vector [22] were engineered, expressing either 4 reprogramming factors (STEMCCA-loxP) or 3 factors plus mCherry (STEMCCA-loxP-RedLight). A loxP site was introduced within the U3 region of the 3' LTR. Upon formation of the provirus a floxed version of each STEMCCA vector is produced. Following exposure to Cre, the entire cassette is excised. LTR: long terminal repeat; PSI: packaging signal; RRE: rev responsive element; cpPu: central polypurine tract; WPRE: Woodchuck hepatitis virus post-transcriptional regulatory element; dU3: deleted U3. (B) Representative images of iPS cells created using the STEMCCA-loxP-RedLight vector, before and after Cre excision. In the top panel (Before Cre), mCherry expression is evident in all iPS colonies, while in the bottom panel (After Cre), red fluorescence expression is no longer detected, illustrating efficient excision of the STEMCCA-loxp-RedLight cassette. (C) Same cells as in (B) were analyzed using flow cytometry to detect mCherry fluorescence. Cells before (red) and after (blue) Cre treatment are shown. (D) Southern blot analysis of genomic DNA (gDNA) purified from 2 representative iPS clones produced with the STEMCCA-loxP vector, before and after Cre-mediated excision. gDNA was digested with BamHI to expose each individual viral integration. Both, SEFL1 and SEFL2 clones displayed a single integration that is not detected after exposure to Cre (SEFL1-Cre and SEFL2-Cre). Similar results were obtained when probing for Klf4. In this case, the endogenous gene was evident in all clones (closed arrowhead) while the STEMCCA encoded Klf4 is not present following Cre excision (open arrowhead). gDNA was digested with BglII to obtain a band of 6.7Kb that confirms appropriate viral transmission. For a control, STEMCCA-loxP plasmid DNA representing 2.5 copies of the insert was digested with BglII. A single band corresponding to an integration of the correct size was observed in SEFL1 and SEFL2 clones and was not present following Cre treatment. (E) Expression of the STEMCCA transcript was analyzed by RT-PCR to confirm excision. As expected, clones SEFL1-Cre and SEFL2-Cre showed no detectable STEMCCA transcript. Nat1 is a constitutively expressed gene and serves as a control for loading.
