Figure 5. Properties of the novel “Coordinator” motif.
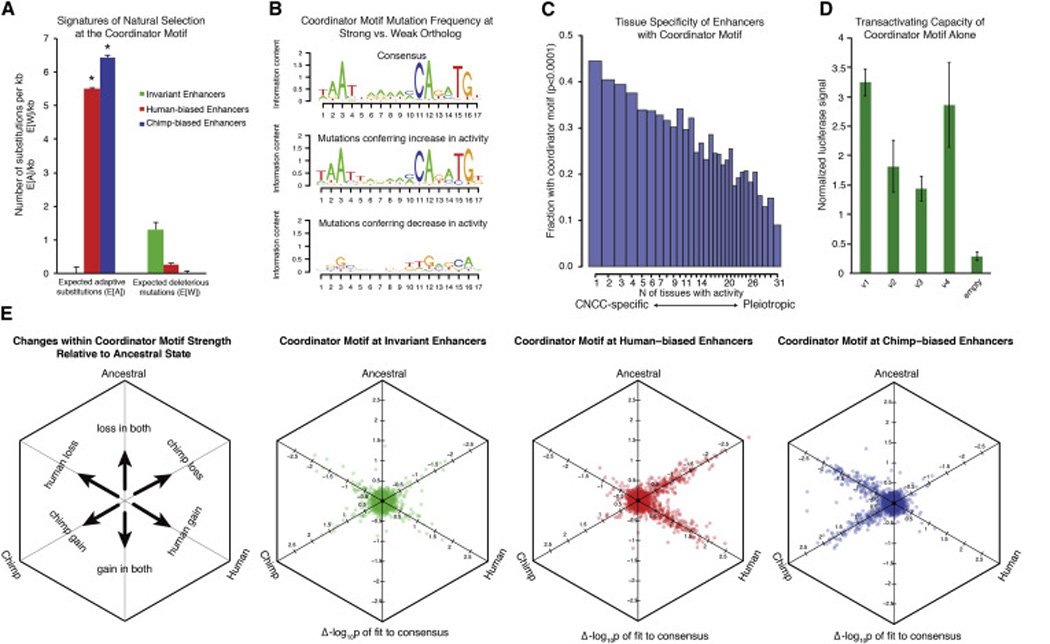
(A) Expected number of adaptive substitutions (E[A]) per kilobase and expected number of deleterious mutations E[W] per kilobase were calculated for all sites of the Coordinator motif at invariant enhancers (green), at human-biased enhancers (red) and at chimp-biased enhancers (blue) using default INSIGHT parameters (Gronau et al., 2013). Significance indicated by * (p<0.01). Overall fractions of nucleotides under selection (ρ) not shown (ρinvariant =0.66, p<0.01; ρhuman-biased = 0.015, p<0.01; ρchimp-biased = 0.019, p<0.01).
(B) Position weight matrix of the Coordinator consensus sequence from top 3000 CNCC specific enhancers is shown (top) relative to logo of mutations preferred at more acetylated (H3K27ac) alleles (middle) versus mutations at less acetylated alleles (bottom).
(C) Enhancers were scored for H3K27ac ChIP-seq enrichments from 30 public data set cell types and binned by number of tissues with activity (1 to 31). The fraction of enhancers per bin with recognizable Coordinator motif (p-value < 0.0001) is indicated on y axis.
(D) Four different versions (V1-4) of the Coordinator motif were cloned in tandem into luciferase reporter vectors and tested for transactivation activity in human CNCCs. Luciferase was normalized relative to renilla transfection control.
(E) Comparison of sequence changes within the Coordinator motif with a reconstructed human-chimp ancestral outgroup. Changes in fit to the Coordinator consensus compared to the ancestral ortholog (−log10p-value) were plotted as orthographic projections along space diagonals for all occurrences of the motif for both human and chimpanzee lineages at different classes of sites. Overlapping data points were offset for better visualization. Schematic shown on the far left.
