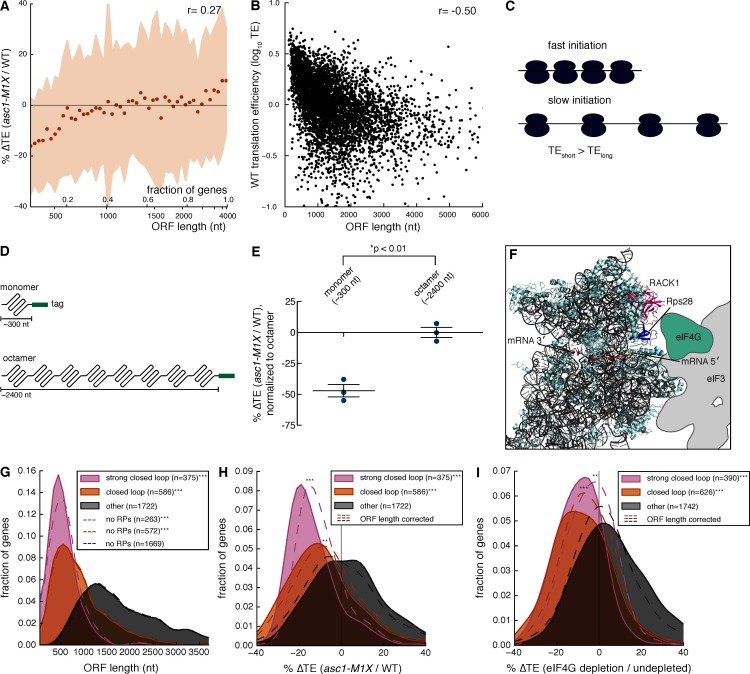
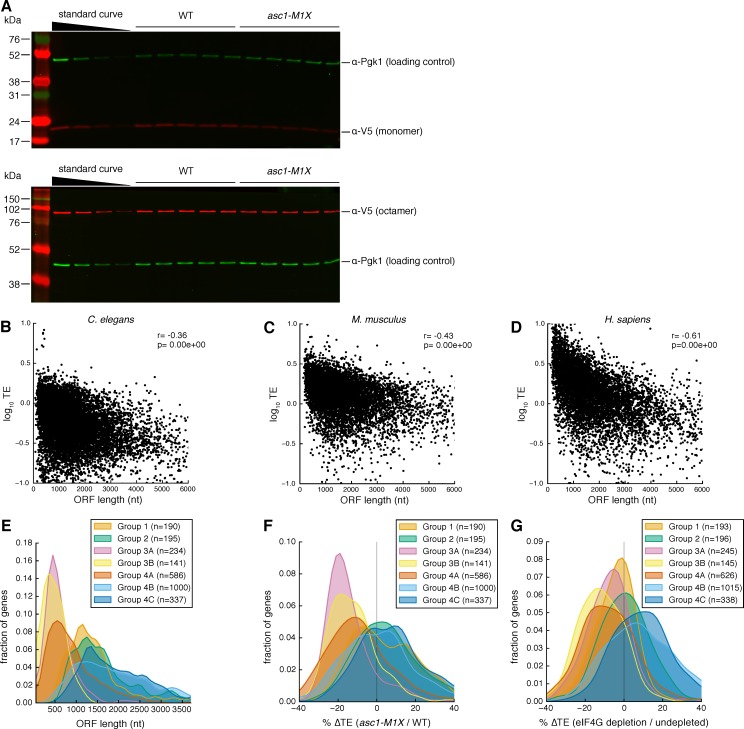
Figure 3. Asc1 is required for efficient translation of short ORFs that form closed loop complexes.
(A) Relationship between ORF length and TE changes in asc1-M1X. The values shown represent the average percent change in TE for bins of 100 genes arranged by length. The ORF lengths shown correspond to the point at which the average ORF length of the bin exceeds the indicated value. Shaded areas represent +/- 1 s.d. from the average change. The ASC1 gene is excluded from the plot. (B) Relationship between ORF length and translational efficiency in WT yeast cells (data from this study). The Spearman correlation coefficient is shown. (C) Model showing the expected effect of a higher initiation rate on short mRNAs compared to long mRNAs on translation efficiency measurements. (D) Diagram of ORF length reporter constructs. The I27 monomer was repeated to make the octamer and each ORF was fused to a C-terminal V5 epitope tag. (E) Result of ORF length reporter experiment. TE is calculated as the normalized protein (V5 tag/Pgk1) to mRNA ratio (V5 mRNA/18S) and the ∆TE (ratio between mutant and WT) is shown. Relative protein concentration was obtained from quantitative Western blotting and mRNA concentration from qRT-PCR. *p=0.002, two-tailed Student’s t-test (monomer vs. octamer). Error bars are SEM from 3 biological replicates derived from independent genetic isolates of asc1-M1X. (F) The structure of the mammalian 48S pre-initiation complex is shown (Lomakin and Steitz, 2013) with the mRNA, RACK1, and Rps28, which crosslinks to the -7 and -10 positions of the mRNA relative to the AUG (Pisarev et al., 2008), indicated. The outline of eIF3 from Hashem et al. (2013) is shown. eIF4G is placed on the left arm of eIF3 based on electron microscopy data from Siridechadilok et al. (2005). (G, H, I) The relationship between closed loop complex association and ORF length (p=10–172and 10–135 for strong closed loop and closed loop groups vs. other mRNAs, respectively) (G), ∆TE in asc1-M1X (p=10–71and 10–42 for strong closed loop and closed loop vs. other mRNAs, respectively) (H), and ∆TE after eIF4G depletion (p=10–70and 10–73 for strong closed loop and closed loop vs. other mRNAs, respectively. Data from Park et al., 2011) (I). In (H) and (I), the dotted lines show the results after accounting for the relationship between ORF length and ∆TE using linear regression. For asc1-M1X, ORF length corrected p-values are 10–30 and 10–14 for strong closed loop and closed loop groups, respectively. For eIF4G depletion, ORF length corrected p-values are 10–17 and 10–28 for strong closed loop and closed loop groups, respectively. p-values are from the one-sided Mann-Whitney U test. Closed loop association groups are from Costello et al. (2015). For G-I, ***p<10–18, **p<10–9, *p<10–3