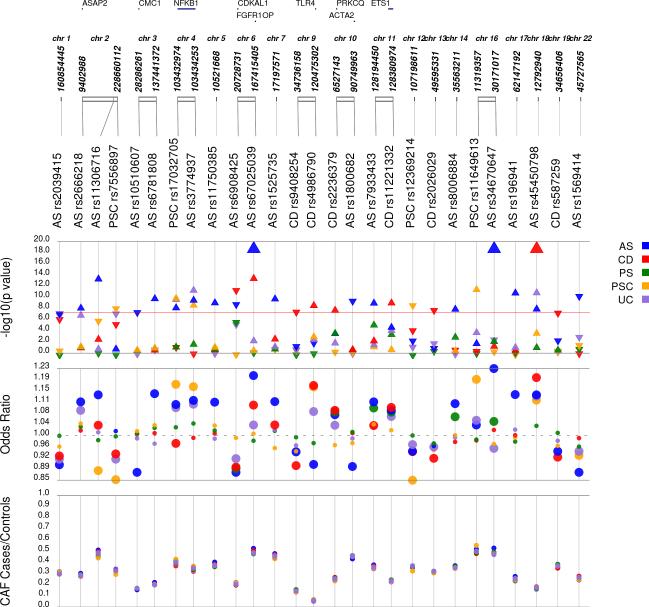
Figure 1. 27 novel genome-wide significant disease associations (Pdisease<5×10−8) for ankylosing spondylitis (AS), Crohn's disease (CD), psoriasis (PS), primary sclerosing cholangitis (PSC) and ulcerative colitis (UC).
Single disease analyses were performed only on SNPs that achieved PSBM<5×10−7 in the primary (unconditioned) cross-disease subset-based association meta-analysis (SBM) approach (see Main Text). We identified 17 novel genome-wide significant susceptibility loci for AS, 6 loci for CD and 4 loci for PSC (Supplementary table 2). Corresponding P-values and ORs for each novel association are shown separately for each disease. With this, the number of known AS, IBD, and PSC risk loci increased to 48, 206, and 20, respectively. For 22 out of 27 gws associations, lead SNPs from the SBM approach (PSBM<5×10−8) and the single disease lookups (PSBM<5×10−7 and Pdisease<5×10−8) are identical, in five instances we have different lead SNPs between SBM and the single disease analyses (Supplementary table 2).
−log10 P-value: −log10 P-values (Pdisease) from Immunochip analysis (Supplementary Table 2) with regard to the physical location of markers; direction of triangle denotes direction of disease-individual effect; OR: odds ratio from the five single disease vs. control subsearches (OR(disease) in Supplementary Table 2). Large circles denote nominal significant disease-individual P-values (Pdisease<0.05); CAF cases/controls: case/control minor allele frequency; If available, the nearest gene within 10kb of the variant is depicted.